Induction of γ-aminobutyric acid plays a positive role to Arabidopsis resistance against Pseudomonas syringae
- PMID: 32458527
- PMCID: PMC7689811
- DOI: 10.1111/jipb.12974
Induction of γ-aminobutyric acid plays a positive role to Arabidopsis resistance against Pseudomonas syringae
Abstract
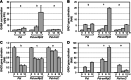
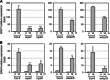
Gamma-aminobutyric acid (GABA) is an important metabolite which functions in plant growth, development, and stress responses. However, its role in plant defense and how it is regulated are largely unknown. Here, we report a detailed analysis of GABA induction during the resistance response to Pseudomonas syringae in Arabidopsis thaliana. While searching for the mechanism underlying the pathogen-responsive mitogen-activated protein kinase (MPK)3/MPK6 signaling cascade in plant immunity, we found that activation of MPK3/MPK6 greatly induced GABA biosynthesis, which is dependent on the glutamate decarboxylase genes GAD1 and GAD4. Inoculation with Pseudomonas syringae pv tomato DC3000 (Pst) and Pst-avrRpt2 expressing the avrRpt2 effector gene induced GAD1 and GAD4 gene expression and increased the levels of GABA. Genetic evidence revealed that GAD1, GAD2, and GAD4 play important roles in both GABA biosynthesis and plant resistance in response to Pst-avrRpt2 infection. The gad1/2/4 triple and gad1/2/4/5 quadruple mutants, in which the GABA levels were extremely low, were more susceptible to both Pst and Pst-avrRpt2. Functional loss of MPK3/MPK6, or their upstream MKK4/MKK5, or their downstream substrate WRKY33 suppressed the induction of GAD1 and GAD4 expression after Pst-avrRpt2 treatment. Our findings shed light on both the regulation and role of GABA in the plant immunity to a bacterial pathogen.
© 2020 The Authors. Journal of Integrative Plant Biology Published by John Wiley & Sons Australia, Ltd on behalf of Institute of Botany, Chinese Academy of Sciences.
Figures





References
-
- Akihiro T, Koike S, Tani R, Tominaga T, Watanabe S, Iijima Y, Aoki K, Shibata D, Ashihara H, Matsukura C, Akama K, Fujimura T, Ezura H (2008) Biochemical mechanism on GABA accumulation during fruit development in tomato. Plant Cell Physiol 49: 1378–1389 - PubMed
-
- Baum G, Chen Y, Arazi T, Takatsuji H, Fromm H (1993) A plant glutamate decarboxylase containing a calmodulin binding domain. Cloning, sequence, and functional analysis. J Biol Chem 268: 19610–19617 - PubMed
-
- Bouché N, Fait A, Zik M, Fromm H (2004) The root‐specific glutamate decarboxylase (GAD1) is essential for sustaining GABA levels in Arabidopsis. Plant Mol Biol 55: 315–325 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

