Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation
- PMID: 32459314
- PMCID: PMC7337904
- DOI: 10.1093/nar/gkaa403
Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation
Abstract
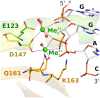
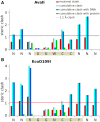
Restriction endonucleases naturally target DNA duplexes. Systematic screening has identified a small minority of these enzymes that can also cleave RNA/DNA heteroduplexes and that may therefore be useful as tools for RNA biochemistry. We have chosen AvaII (G↓GWCC, where W stands for A or T) as a representative of this group of restriction endonucleases for detailed characterization. Here, we report crystal structures of AvaII alone, in specific complex with partially cleaved dsDNA, and in scanning complex with an RNA/DNA hybrid. The specific complex reveals a novel form of semi-specific dsDNA readout by a hexa-coordinated metal cation, most likely Ca2+ or Mg2+. Substitutions of residues anchoring this non-catalytic metal ion severely impair DNA binding and cleavage. The dsDNA in the AvaII complex is in the A-like form. This creates space for 2'-OH groups to be accommodated without intra-nucleic acid steric conflicts. PD-(D/E)XK restriction endonucleases of known structure that bind their dsDNA targets in the A-like form cluster into structurally similar groups. Most such enzymes, including some not previously studied in this respect, cleave RNA/DNA heteroduplexes. We conclude that A-form dsDNA binding is a good predictor for RNA/DNA cleavage activity.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

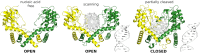
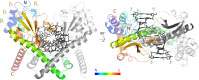




References
-
- Arnott S. Principles of Nucleic-Acid Structure—Saenger,W. Nature. 1984; 312:174–174.
-
- Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A.. Structural basis for double-stranded RNA processing by Dicer. Science. 2006; 311:195–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

