Genetic diversity of influenza A viruses circulating in Bulgaria during the 2018-2019 winter season
- PMID: 32459617
- PMCID: PMC7481746
- DOI: 10.1099/jmm.0.001198
Genetic diversity of influenza A viruses circulating in Bulgaria during the 2018-2019 winter season
Abstract
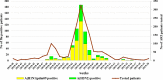
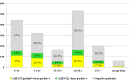
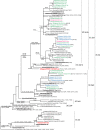
Introduction. Influenza viruses evolve rapidly and change their antigenic characteristics, necessitating biannual updates of flu vaccines.Aim. The aim of this study was to characterize influenza viruses circulating in Bulgaria during the 2018/2019 season and to identify amino acid substitutions in them that might impact vaccine effectiveness.Methodology. Typing/subtyping of influenza viruses were performed using real-time Reverse Transcription-PCR (RT-PCR) and results of phylogenetic and amino acid sequence analyses of influenza strains are presented.Results. A(H1N1)pdm09 (66 %) predominated over A(H3N2) (34 %) viruses, with undetected circulation of B viruses in the 2018/2019 season. All A(H1N1)pdm09 viruses studied fell into the recently designated 6B.1A subclade with over 50 % falling in four subgroups: 6B.1A2, 6B.1A5, 6B.1A6 and 6B.1A7. Analysed A(H3N2) viruses belonged to subclades 3C.2a1b and 3C.2a2. Amino acid sequence analysis of 36 A(H1N1)pdm09 isolates revealed the presence of six-ten substitutions in haemagglutinin (HA), compared to the A/Michigan/45/2015 vaccine virus, three of which occurred in antigenic sites Sa and Cb, together with four-nine changes at positions in neuraminidase (NA), and a number of substitutions in internal proteins. HA1 D222N substitution, associated with increased virulence, was identified in two A(H1N1)pdm09 viruses. Despite the presence of several amino acid substitutions, A(H1N1)pdm09 viruses remained antigenically similar to the vaccine virus. The 28 A(H3N2) viruses characterized carried substitutions in HA, including some in antigenic sites A, B, C and E, in NA and internal protein sequences.Conclusion. The results of this study showed the genetic diversity of circulating influenza viruses and the need for continuous antigenic and molecular surveillance.
Keywords: amino acid substitution; genetic characterisation; influenza virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- WHO Influenza fact sheet. influenza (seasonal) fact sheet. 6 November 2018; available at. 2018a https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

