Extraction of Information Related to Drug Safety Surveillance From Electronic Health Record Notes: Joint Modeling of Entities and Relations Using Knowledge-Aware Neural Attentive Models
- PMID: 32459650
- PMCID: PMC7382020
- DOI: 10.2196/18417
Extraction of Information Related to Drug Safety Surveillance From Electronic Health Record Notes: Joint Modeling of Entities and Relations Using Knowledge-Aware Neural Attentive Models
Abstract
Background: An adverse drug event (ADE) is commonly defined as "an injury resulting from medical intervention related to a drug." Providing information related to ADEs and alerting caregivers at the point of care can reduce the risk of prescription and diagnostic errors and improve health outcomes. ADEs captured in structured data in electronic health records (EHRs) as either coded problems or allergies are often incomplete, leading to underreporting. Therefore, it is important to develop capabilities to process unstructured EHR data in the form of clinical notes, which contain a richer documentation of a patient's ADE. Several natural language processing (NLP) systems have been proposed to automatically extract information related to ADEs. However, the results from these systems showed that significant improvement is still required for the automatic extraction of ADEs from clinical notes.
Objective: This study aims to improve the automatic extraction of ADEs and related information such as drugs, their attributes, and reason for administration from the clinical notes of patients.
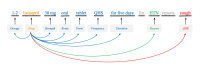
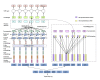
Methods: This research was conducted using discharge summaries from the Medical Information Mart for Intensive Care III (MIMIC-III) database obtained through the 2018 National NLP Clinical Challenges (n2c2) annotated with drugs, drug attributes (ie, strength, form, frequency, route, dosage, duration), ADEs, reasons, and relations between drugs and other entities. We developed a deep learning-based system for extracting these drug-centric concepts and relations simultaneously using a joint method enhanced with contextualized embeddings, a position-attention mechanism, and knowledge representations. The joint method generated different sentence representations for each drug, which were then used to extract related concepts and relations simultaneously. Contextualized representations trained on the MIMIC-III database were used to capture context-sensitive meanings of words. The position-attention mechanism amplified the benefits of the joint method by generating sentence representations that capture long-distance relations. Knowledge representations were obtained from graph embeddings created using the US Food and Drug Administration Adverse Event Reporting System database to improve relation extraction, especially when contextual clues were insufficient.
Results: Our system achieved new state-of-the-art results on the n2c2 data set, with significant improvements in recognizing crucial drug-reason (F1=0.650 versus F1=0.579) and drug-ADE (F1=0.490 versus F1=0.476) relations.
Conclusions: This study presents a system for extracting drug-centric concepts and relations that outperformed current state-of-the-art results and shows that contextualized embeddings, position-attention mechanisms, and knowledge graph embeddings effectively improve deep learning-based concepts and relation extraction. This study demonstrates the potential for deep learning-based methods to help extract real-world evidence from unstructured patient data for drug safety surveillance.
Keywords: adverse drug events; adverse drug reaction reporting systems; deep learning; electronic health records; information extraction; named entity recognition; natural language processing; relation extraction.
©Bharath Dandala, Venkata Joopudi, Ching-Huei Tsou, Jennifer J Liang, Parthasarathy Suryanarayanan. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 10.07.2020.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures



Similar articles
-
Extraction of Information Related to Adverse Drug Events from Electronic Health Record Notes: Design of an End-to-End Model Based on Deep Learning.JMIR Med Inform. 2018 Nov 26;6(4):e12159. doi: 10.2196/12159. JMIR Med Inform. 2018. PMID: 30478023 Free PMC article.
-
Clinical Relation Extraction Toward Drug Safety Surveillance Using Electronic Health Record Narratives: Classical Learning Versus Deep Learning.JMIR Public Health Surveill. 2018 Apr 25;4(2):e29. doi: 10.2196/publichealth.9361. JMIR Public Health Surveill. 2018. PMID: 29695376 Free PMC article.
-
A Hybrid Model for Family History Information Identification and Relation Extraction: Development and Evaluation of an End-to-End Information Extraction System.JMIR Med Inform. 2021 Apr 22;9(4):e22797. doi: 10.2196/22797. JMIR Med Inform. 2021. PMID: 33885370 Free PMC article.
-
Overview of the First Natural Language Processing Challenge for Extracting Medication, Indication, and Adverse Drug Events from Electronic Health Record Notes (MADE 1.0).Drug Saf. 2019 Jan;42(1):99-111. doi: 10.1007/s40264-018-0762-z. Drug Saf. 2019. PMID: 30649735 Free PMC article. Review.
-
Natural Language Processing of Clinical Notes on Chronic Diseases: Systematic Review.JMIR Med Inform. 2019 Apr 27;7(2):e12239. doi: 10.2196/12239. JMIR Med Inform. 2019. PMID: 31066697 Free PMC article. Review.
Cited by
-
Detecting Adverse Drug Events Through the Chronological Relationship Between the Medication Period and the Presence of Adverse Reactions From Electronic Medical Record Systems: Observational Study.JMIR Med Inform. 2021 Nov 1;9(11):e28763. doi: 10.2196/28763. JMIR Med Inform. 2021. PMID: 33993103 Free PMC article.
-
Extracting medication changes in clinical narratives using pre-trained language models.J Biomed Inform. 2023 Mar;139:104302. doi: 10.1016/j.jbi.2023.104302. Epub 2023 Feb 6. J Biomed Inform. 2023. PMID: 36754129 Free PMC article.
-
Health Care Language Models and Their Fine-Tuning for Information Extraction: Scoping Review.JMIR Med Inform. 2024 Oct 21;12:e60164. doi: 10.2196/60164. JMIR Med Inform. 2024. PMID: 39432345 Free PMC article.
-
Adverse drug event detection using natural language processing: A scoping review of supervised learning methods.PLoS One. 2023 Jan 3;18(1):e0279842. doi: 10.1371/journal.pone.0279842. eCollection 2023. PLoS One. 2023. PMID: 36595517 Free PMC article.
References
-
- Gunter TD, Terry NP. The emergence of national electronic health record architectures in the United States and Australia: models, costs, and questions. J Med Internet Res. 2005 Mar 14;7(1):e3. doi: 10.2196/jmir.7.1.e3. https://www.jmir.org/2005/1/e3/ - DOI - PMC - PubMed
-
- Rosenbloom S, Stead W, Denny J, Giuse D, Lorenzi N, Brown S, Johnson K. Generating clinical notes for electronic health record systems. Appl Clin Inform. 2010 Jan 1;1(3):232–43. doi: 10.4338/ACI-2010-03-RA-0019. http://europepmc.org/abstract/MED/21031148 - DOI - PMC - PubMed
-
- Bates DW, Cullen DJ, Laird N, Petersen LA, Small SD, Servi D, Laffel G, Sweitzer BJ, Shea BF, Hallisey R. Incidence of adverse drug events and potential adverse drug events. Implications for prevention. ADE prevention study group. J Am Med Assoc. 1995 Jul 5;274(1):29–34. - PubMed
-
- Johnson JA, Bootman JL. Drug-related morbidity and mortality. A cost-of-illness model. Arch Intern Med. 1995 Oct 9;155(18):1949–56. - PubMed
-
- Classen DC, Pestotnik SL, Evans RS, Lloyd JF, Burke JP. Adverse drug events in hospitalized patients. Excess length of stay, extra costs, and attributable mortality. J Am Med Assoc. 1997;277(4):301–6. - PubMed
LinkOut - more resources
Full Text Sources

