Expression of RUNX1-ETO Rapidly Alters the Chromatin Landscape and Growth of Early Human Myeloid Precursor Cells
- PMID: 32460028
- PMCID: PMC7262600
- DOI: 10.1016/j.celrep.2020.107691
Expression of RUNX1-ETO Rapidly Alters the Chromatin Landscape and Growth of Early Human Myeloid Precursor Cells
Abstract
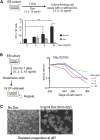
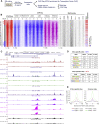
Acute myeloid leukemia (AML) is a hematopoietic malignancy caused by recurrent mutations in genes encoding transcriptional, chromatin, and/or signaling regulators. The t(8;21) translocation generates the aberrant transcription factor RUNX1-ETO (RUNX1-RUNX1T1), which by itself is insufficient to cause disease. t(8;21) AML patients show extensive chromatin reprogramming and have acquired additional mutations. Therefore, the genomic and developmental effects directly and solely attributable to RUNX1-ETO expression are unclear. To address this, we employ a human embryonic stem cell differentiation system capable of forming definitive myeloid progenitor cells to express RUNX1-ETO in an inducible fashion. Induction of RUNX1-ETO causes extensive chromatin reprogramming by interfering with RUNX1 binding, blocks differentiation, and arrests cellular growth, whereby growth arrest is reversible following RUNX1-ETO removal. Single-cell gene expression analyses show that RUNX1-ETO induction alters the differentiation of early myeloid progenitors, but not of other progenitor types, indicating that oncoprotein-mediated transcriptional reprogramming is highly target cell specific.
Keywords: Acute Myeloid Leukemia (AML); RUNX1-ETO; chromatin; human ES cell differentiation; myelopoiesis; single cell RNA-Seq.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures






References
-
- Ben-Ami O., Friedman D., Leshkowitz D., Goldenberg D., Orlovsky K., Pencovich N., Lotem J., Tanay A., Groner Y. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013;4:1131–1143. - PubMed
-
- Boisset J.-C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

