SPOP promotes ubiquitination and degradation of LATS1 to enhance kidney cancer progression
- PMID: 32460168
- PMCID: PMC7248661
- DOI: 10.1016/j.ebiom.2020.102795
SPOP promotes ubiquitination and degradation of LATS1 to enhance kidney cancer progression
Abstract
Background: Emerging evidence has demonstrated that SPOP functions as an oncoprotein in kidney cancer to promote tumorigenesis by ubiquitination-mediated degradation of multiple regulators of cellular proliferation and apoptosis. However, the detailed molecular mechanism underlying the oncogenic role of SPOP in kidney tumorigenesis remains elusive.
Methods: Multiple approaches such as Co-IP, Transfection, RT-PCR, Western blotting, and animal studies were utilized to explore the role of SPOP in kidney cancer.
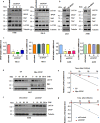
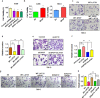
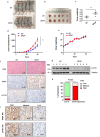
Findings: Here we identified LATS1, a critical component of the Hippo tumour suppressor pathway, as a novel ubiquitin substrate of SPOP. We found that LATS1 interacted with Cullin3, and depletion of Cullin 3 upregulated the abundance of LATS1 largely via prolonging LATS1 protein half-life. Mechanistically, SPOP specifically interacted with LATS1, and promoted the poly-ubiquitination and subsequent degradation of LATS1 in a degron-dependent manner. As such, over-expression of SPOP promoted cell proliferation partly through regulating cell cycle distribution in kidney cancer cells. Furthermore, SPOP also promoted kidney cancer cell invasion via degrading LATS1.
Interpretation: Our study provides evidence for a novel mechanism of SPOP in kidney cancer progression in part through promoting degradation of the LATS1 tumour suppressor.
Keywords: Degradation; Growth; Kidney; LATS1; SPOP; Ubiquitination.
Copyright © 2020 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no conflict of interest.
Figures



Comment in
-
Taking a SPOP at renal cell carcinoma - unraveling a novel pathway for Tumor progression in clear cell RCC.EBioMedicine. 2020 Jun;56:102823. doi: 10.1016/j.ebiom.2020.102823. Epub 2020 Jun 5. EBioMedicine. 2020. PMID: 32512506 Free PMC article. No abstract available.
Similar articles
-
Prostate cancer-associated mutation in SPOP impairs its ability to target Cdc20 for poly-ubiquitination and degradation.Cancer Lett. 2017 Jan 28;385:207-214. doi: 10.1016/j.canlet.2016.10.021. Epub 2016 Oct 22. Cancer Lett. 2017. PMID: 27780719 Free PMC article.
-
Cullin 3SPOP ubiquitin E3 ligase promotes the poly-ubiquitination and degradation of HDAC6.Oncotarget. 2017 Jul 18;8(29):47890-47901. doi: 10.18632/oncotarget.18141. Oncotarget. 2017. PMID: 28599312 Free PMC article.
-
SPOP promotes CDCA5 degradation to regulate prostate cancer progression via the AKT pathway.Neoplasia. 2021 Oct;23(10):1037-1047. doi: 10.1016/j.neo.2021.08.002. Epub 2021 Sep 10. Neoplasia. 2021. PMID: 34509929 Free PMC article.
-
The emerging role of SPOP protein in tumorigenesis and cancer therapy.Mol Cancer. 2020 Jan 4;19(1):2. doi: 10.1186/s12943-019-1124-x. Mol Cancer. 2020. PMID: 31901237 Free PMC article. Review.
-
Novel insights into the SPOP E3 ubiquitin ligase: From the regulation of molecular mechanisms to tumorigenesis.Biomed Pharmacother. 2022 May;149:112882. doi: 10.1016/j.biopha.2022.112882. Epub 2022 Mar 29. Biomed Pharmacother. 2022. PMID: 35364375 Review.
Cited by
-
N6-methyladenosine-modified DBT alleviates lipid accumulation and inhibits tumor progression in clear cell renal cell carcinoma through the ANXA2/YAP axis-regulated Hippo pathway.Cancer Commun (Lond). 2023 Apr;43(4):480-502. doi: 10.1002/cac2.12413. Epub 2023 Mar 1. Cancer Commun (Lond). 2023. PMID: 36860124 Free PMC article.
-
Degradation of DRAK1 by CUL3/SPOP E3 Ubiquitin ligase promotes tumor growth of paclitaxel-resistant cervical cancer cells.Cell Death Dis. 2022 Feb 22;13(2):169. doi: 10.1038/s41419-022-04619-w. Cell Death Dis. 2022. PMID: 35194034 Free PMC article.
-
Nosip is a potential therapeutic target in hepatocellular carcinoma cells.iScience. 2023 Jul 10;26(8):107353. doi: 10.1016/j.isci.2023.107353. eCollection 2023 Aug 18. iScience. 2023. PMID: 37529099 Free PMC article.
-
Taking a SPOP at renal cell carcinoma - unraveling a novel pathway for Tumor progression in clear cell RCC.EBioMedicine. 2020 Jun;56:102823. doi: 10.1016/j.ebiom.2020.102823. Epub 2020 Jun 5. EBioMedicine. 2020. PMID: 32512506 Free PMC article. No abstract available.
-
Challenges and opportunities for the diverse substrates of SPOP E3 ubiquitin ligase in cancer.Theranostics. 2025 May 8;15(13):6111-6145. doi: 10.7150/thno.113356. eCollection 2025. Theranostics. 2025. PMID: 40521202 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Lopez-Beltran A, Scarpelli M, Montironi R. WHO classification of the renal tumors of the adults. Eur Urol. 2006;49(5):798–805. - PubMed
-
- Hollingsworth JM, Miller DC, Daignault S. Five-year survival after surgical treatment for kidney cancer: a population-based competing risk analysis. Cancer. 2007;109(9):1763–1768. - PubMed
-
- Interferon-alpha and survival in metastatic renal carcinoma: early results of a randomised controlled trial. Med Res Council Renal Cancer Collab Lancet. 1999;353(9146):14–17. - PubMed
-
- Pyrhonen S, Salminen E, Ruutu M. Prospective randomized trial of interferon alfa-2a plus vinblastine versus vinblastine alone in patients with advanced renal cell cancer. J Clin Oncol. 1999;17(9):2859–2867. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

