FOXD1-AS1 regulates FOXD1 translation and promotes gastric cancer progression and chemoresistance by activating the PI3K/AKT/mTOR pathway
- PMID: 32460412
- PMCID: PMC7782086
- DOI: 10.1002/1878-0261.12728
FOXD1-AS1 regulates FOXD1 translation and promotes gastric cancer progression and chemoresistance by activating the PI3K/AKT/mTOR pathway
Abstract
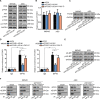
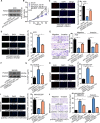
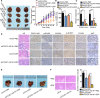
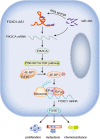
Gastric cancer (GC) is a common gastrointestinal cancer with a high global mortality. Recent reports have suggested that long noncoding RNA (lncRNA) are implicated in multiple aspects of GC, including pathogenesis, progression, and therapeutic response. Herein, we investigated the function of FOXD1-AS1 in GC progression and chemoresistance. Expression of FOXD1-AS1 was low in normal stomach tissues but was upregulated in GC cell lines. Silencing of FOXD1-AS1 impaired GC cell proliferation and motility in vitro, and repressed tumor growth and metastasis in vivo. Importantly, FOXD1-AS1 upregulation increased the resistance of GC cells to cisplatin. Moreover, we found that FOXD1-AS1 promoted FOXD1 protein translation through the eIF4G-eIF4E-eIF4A translational complex. We also demonstrated that FOXD1-AS1 released eIF4E from phosphorylated 4E-BP1 and thereby strengthened the interaction of eIF4E with eIF4G by activating the PI3K/AKT/mTOR pathway. Activation of the PI3K/AKT/mTOR pathway was due to the post-transcriptional upregulation of PIK3CA, in turn induced by FOXD1-AS1-mediated sequestering of microRNA (miR)-466. Furthermore, we verified that FOXD1-AS1 facilitated GC progression and cisplatin resistance in a FOXD1-dependent manner. In conclusion, FOXD1-AS1 aggravates GC progression and chemoresistance by promoting FOXD1 translation via PIK3CA/PI3K/AKT/mTOR signaling. These findings highlight a novel target for treatment of patients GC, particularly patients with cisplatin resistance.
Keywords: FOXD1; FOXD1-AS1; PI3K/AKT/mTOR pathway; eIF4G-eIF4E-eIF4A translational complex; gastric cancer.
© 2020 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- (2015) Cancer cells require eIF4E to translate pro‐oncogenic mRNAs. Cancer Discov 5, 792.
-
- Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, Chen Q, Wei C, Fu H, Xu T et al (2018) Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR‐204‐5p in gastric cancer. Clin Cancer Res 24, 2002–2014. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

