Single-cell RNA sequencing reveals the tumor microenvironment and facilitates strategic choices to circumvent treatment failure in a chemorefractory bladder cancer patient
- PMID: 32460812
- PMCID: PMC7251908
- DOI: 10.1186/s13073-020-00741-6
Single-cell RNA sequencing reveals the tumor microenvironment and facilitates strategic choices to circumvent treatment failure in a chemorefractory bladder cancer patient
Abstract
Background: Tumor cell-intrinsic mechanisms and complex interactions with the tumor microenvironment contribute to therapeutic failure via tumor evolution. It may be possible to overcome treatment resistance by developing a personalized approach against relapsing cancers based on a comprehensive analysis of cell type-specific transcriptomic changes over the clinical course of the disease using single-cell RNA sequencing (scRNA-seq).
Methods: Here, we used scRNA-seq to depict the tumor landscape of a single case of chemo-resistant metastatic, muscle-invasive urothelial bladder cancer (MIUBC) addicted to an activating Harvey rat sarcoma viral oncogene homolog (HRAS) mutation. In order to analyze tumor evolution and microenvironmental changes upon treatment, we also applied scRNA-seq to the corresponding patient-derived xenograft (PDX) before and after treatment with tipifarnib, a HRAS-targeting agent under clinical evaluation.
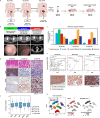
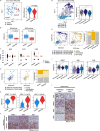
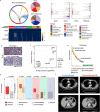
Results: In the parallel analysis of the human MIUBC and the PDX, diverse stromal and immune cell populations recapitulated the cellular composition in the human and mouse tumor microenvironment. Treatment with tipifarnib showed dramatic anticancer effects but was unable to achieve a complete response. Importantly, the comparative scRNA-seq analysis between pre- and post-tipifarnib-treated PDX revealed the nature of tipifarnib-refractory tumor cells and the tumor-supporting microenvironment. Based on the upregulation of programmed death-ligand 1 (PD-L1) in surviving tumor cells, and the accumulation of multiple immune-suppressive subsets from post-tipifarnib-treated PDX, a PD-L1 inhibitor, atezolizumab, was clinically applied; this resulted in a favorable response from the patient with acquired resistance to tipifarnib.
Conclusion: We presented a single case report demonstrating the power of scRNA-seq for visualizing the tumor microenvironment and identifying molecular and cellular therapeutic targets in a treatment-refractory cancer patient.
Keywords: Muscle-invasive urothelial bladder cancer; Single-cell RNA sequencing; Treatment resistance; Tumor microenvironment; Tumoral heterogeneity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Dzobo K, Senthebane DA, Thomford NE, Rowe A, Dandara C, Parker MI. Not everyone fits the mold: intratumor and intertumor heterogeneity and innovative cancer drug design and development. OMICS. 2018;22:17–34. - PubMed
-
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

