Semisynthesis of an evasin from tick saliva reveals a critical role of tyrosine sulfation for chemokine binding and inhibition
- PMID: 32461364
- PMCID: PMC7293604
- DOI: 10.1073/pnas.2000605117
Semisynthesis of an evasin from tick saliva reveals a critical role of tyrosine sulfation for chemokine binding and inhibition
Abstract
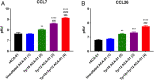
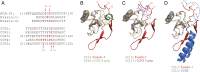
Blood-feeding arthropods produce antiinflammatory salivary proteins called evasins that function through inhibition of chemokine-receptor signaling in the host. Herein, we show that the evasin ACA-01 from the Amblyomma cajennense tick can be posttranslationally sulfated at two tyrosine residues, albeit as a mixture of sulfated variants. Homogenously sulfated variants of the proteins were efficiently assembled via a semisynthetic native chemical ligation strategy. Sulfation significantly improved the binding affinity of ACA-01 for a range of proinflammatory chemokines and enhanced the ability of ACA-01 to inhibit chemokine signaling through cognate receptors. Comparisons of evasin sequences and structural data suggest that tyrosine sulfation serves as a receptor mimetic strategy for recognizing and suppressing the proinflammatory activity of a wide variety of mammalian chemokines. As such, the incorporation of this posttranslational modification (PTM) or mimics thereof into evasins may provide a strategy to optimize tick salivary proteins for antiinflammatory applications.
Keywords: antiinflammatory; chemokines; evasins; sulfation; ticks.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Bonecchi R. et al., Chemokines and chemokine receptors: An overview. Front. Biosci. 14, 540–551 (2009). - PubMed
-
- Rollins B. J., Chemokines. Blood 90, 909–928 (1997). - PubMed
-
- Charo I. F., Peters W., Chemokine receptor 2 (CCR2) in atherosclerosis, infectious diseases, and regulation of T-cell polarization. Microcirculation 10, 259–264 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

