Photothermal modulation of human stem cells using light-responsive 2D nanomaterials
- PMID: 32461372
- PMCID: PMC7306823
- DOI: 10.1073/pnas.1914345117
Photothermal modulation of human stem cells using light-responsive 2D nanomaterials
Abstract
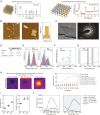
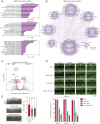
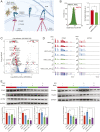
Two-dimensional (2D) molybdenum disulfide (MoS2) nanomaterials are an emerging class of biomaterials that are photoresponsive at near-infrared wavelengths (NIR). Here, we demonstrate the ability of 2D MoS2 to modulate cellular functions of human stem cells through photothermal mechanisms. The interaction of MoS2 and NIR stimulation of MoS2 with human stem cells is investigated using whole-transcriptome sequencing (RNA-seq). Global gene expression profile of stem cells reveals significant influence of MoS2 and NIR stimulation of MoS2 on integrins, cellular migration, and wound healing. The combination of MoS2 and NIR light may provide new approaches to regulate and direct these cellular functions for the purposes of regenerative medicine as well as cancer therapy.
Keywords: cell adhesion; cell–nanoparticles interactions; human mesenchymal stem cells; two-dimensional (2D) nanomaterials; whole-transcriptome sequencing.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Novoselov K. S., Mishchenko A., Carvalho A., Castro Neto A. H., 2D materials and van der Waals heterostructures. Science 353, aac9439 (2016). - PubMed
-
- Chimene D., Alge D. L., Gaharwar A. K., Two-dimensional nanomaterials for biomedical applications: Emerging trends and future prospects. Adv. Mater. 27, 7261–7284 (2015). - PubMed
-
- Chhowalla M. et al., The chemistry of two-dimensional layered transition metal dichalcogenide nanosheets. Nat. Chem. 5, 263–275 (2013). - PubMed
-
- Liu T. et al., Drug delivery with PEGylated MoS2 nano-sheets for combined photothermal and chemotherapy of cancer. Adv. Mater. 26, 3433–3440 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

