The DNA methylation landscape of giant viruses
- PMID: 32461636
- PMCID: PMC7253447
- DOI: 10.1038/s41467-020-16414-2
The DNA methylation landscape of giant viruses
Abstract
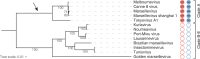
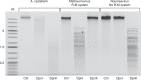
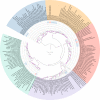
DNA methylation is an important epigenetic mark that contributes to various regulations in all domains of life. Giant viruses are widespread dsDNA viruses with gene contents overlapping the cellular world that also encode DNA methyltransferases. Yet, virtually nothing is known about the methylation of their DNA. Here, we use single-molecule real-time sequencing to study the complete methylome of a large spectrum of giant viruses. We show that DNA methylation is widespread, affecting 2/3 of the tested families, although unevenly distributed. We also identify the corresponding viral methyltransferases and show that they are subject to intricate gene transfers between bacteria, viruses and their eukaryotic host. Most methyltransferases are conserved, functional and under purifying selection, suggesting that they increase the viruses' fitness. Some virally encoded methyltransferases are also paired with restriction endonucleases forming Restriction-Modification systems. Our data suggest that giant viruses' methyltransferases are involved in diverse forms of virus-pathogens interactions during coinfections.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Evolution of Restriction-Modification Systems Consisting of One Restriction Endonuclease and Two DNA Methyltransferases.Biochemistry (Mosc). 2023 Feb;88(2):253-261. doi: 10.1134/S0006297923020086. Biochemistry (Mosc). 2023. PMID: 37072330
-
Evolution of ubiquitin, cytoskeleton, and vesicular trafficking machinery in giant viruses.J Virol. 2025 Mar 18;99(3):e0171524. doi: 10.1128/jvi.01715-24. Epub 2025 Feb 11. J Virol. 2025. PMID: 39932282 Free PMC article.
-
Evolution of the Large Nucleocytoplasmic DNA Viruses of Eukaryotes and Convergent Origins of Viral Gigantism.Adv Virus Res. 2019;103:167-202. doi: 10.1016/bs.aivir.2018.09.002. Epub 2018 Nov 10. Adv Virus Res. 2019. PMID: 30635076 Review.
-
A positive perspective on DNA methylation: regulatory functions of DNA methylation outside of host defense in Gram-positive bacteria.Crit Rev Biochem Mol Biol. 2020 Dec;55(6):576-591. doi: 10.1080/10409238.2020.1828257. Epub 2020 Oct 15. Crit Rev Biochem Mol Biol. 2020. PMID: 33059472
-
Giant viruses and their mobile genetic elements: the molecular symbiosis hypothesis.Curr Opin Virol. 2018 Dec;33:81-88. doi: 10.1016/j.coviro.2018.07.013. Epub 2018 Aug 13. Curr Opin Virol. 2018. PMID: 30114664 Review.
Cited by
-
Metagenomic characterization of viruses and mobile genetic elements associated with the DPANN archaeal superphylum.Nat Microbiol. 2024 Dec;9(12):3362-3375. doi: 10.1038/s41564-024-01839-y. Epub 2024 Oct 24. Nat Microbiol. 2024. PMID: 39448846
-
Long-Read Sequencing Reveals Extensive DNA Methylations in Human Gut Phagenome Contributed by Prevalently Phage-Encoded Methyltransferases.Adv Sci (Weinh). 2023 Sep;10(25):e2302159. doi: 10.1002/advs.202302159. Epub 2023 Jun 29. Adv Sci (Weinh). 2023. PMID: 37382405 Free PMC article.
-
PacBio sequencing of human fecal samples uncovers the DNA methylation landscape of 22 673 gut phages.Nucleic Acids Res. 2023 Dec 11;51(22):12140-12149. doi: 10.1093/nar/gkad977. Nucleic Acids Res. 2023. PMID: 37904586 Free PMC article.
-
Unveiling Prasinovirus diversity and host specificity through targeted enrichment in the South China Sea.ISME Commun. 2024 Aug 29;4(1):ycae109. doi: 10.1093/ismeco/ycae109. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39296779 Free PMC article.
-
Diagnosis and monitoring of virus-associated cancer using cell-free DNA.Curr Opin Virol. 2023 Jun;60:101331. doi: 10.1016/j.coviro.2023.101331. Epub 2023 May 13. Curr Opin Virol. 2023. PMID: 37187125 Free PMC article. Review.
References
-
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. - PubMed
-
- Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. - PubMed
-
- Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

