The stepwise assembly of the neonatal virome is modulated by breastfeeding
- PMID: 32461640
- PMCID: PMC7263352
- DOI: 10.1038/s41586-020-2192-1
The stepwise assembly of the neonatal virome is modulated by breastfeeding
Abstract
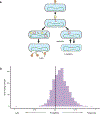
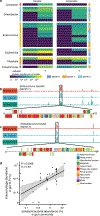
The gut of healthy human neonates is usually devoid of viruses at birth, but quickly becomes colonized, which-in some cases-leads to gastrointestinal disorders1-4. Here we show that the assembly of the viral community in neonates takes place in distinct steps. Fluorescent staining of virus-like particles purified from infant meconium or early stool samples shows few or no particles, but by one month of life particle numbers increase to 109 per gram, and these numbers seem to persist throughout life5-7. We investigated the origin of these viral populations using shotgun metagenomic sequencing of virus-enriched preparations and whole microbial communities, followed by targeted microbiological analyses. Results indicate that, early after birth, pioneer bacteria colonize the infant gut and by one month prophages induced from these bacteria provide the predominant population of virus-like particles. By four months of life, identifiable viruses that replicate in human cells become more prominent. Multiple human viruses were more abundant in stool samples from babies who were exclusively fed on formula milk compared with those fed partially or fully on breast milk, paralleling reports that breast milk can be protective against viral infections8-10. Bacteriophage populations also differed depending on whether or not the infant was breastfed. We show that the colonization of the infant gut is stepwise, first mainly by temperate bacteriophages induced from pioneer bacteria, and later by viruses that replicate in human cells; this second phase is modulated by breastfeeding.
Figures



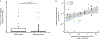
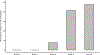

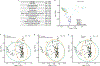




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

