Origin of complexity in haemoglobin evolution
- PMID: 32461643
- PMCID: PMC8259614
- DOI: 10.1038/s41586-020-2292-y
Origin of complexity in haemoglobin evolution
Erratum in
-
Author Correction: Origin of complexity in haemoglobin evolution.Nature. 2020 Jul;583(7816):E26. doi: 10.1038/s41586-020-2472-9. Nature. 2020. PMID: 32587402
Abstract
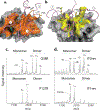
Most proteins associate into multimeric complexes with specific architectures1,2, which often have functional properties such as cooperative ligand binding or allosteric regulation3. No detailed knowledge is available about how any multimer and its functions arose during evolution. Here we use ancestral protein reconstruction and biophysical assays to elucidate the origins of vertebrate haemoglobin, a heterotetramer of paralogous α- and β-subunits that mediates respiratory oxygen transport and exchange by cooperatively binding oxygen with moderate affinity. We show that modern haemoglobin evolved from an ancient monomer and characterize the historical 'missing link' through which the modern tetramer evolved-a noncooperative homodimer with high oxygen affinity that existed before the gene duplication that generated distinct α- and β-subunits. Reintroducing just two post-duplication historical substitutions into the ancestral protein is sufficient to cause strong tetramerization by creating favourable contacts with more ancient residues on the opposing subunit. These surface substitutions markedly reduce oxygen affinity and even confer cooperativity, because an ancient linkage between the oxygen binding site and the multimerization interface was already an intrinsic feature of the protein's structure. Our findings establish that evolution can produce new complex molecular structures and functions via simple genetic mechanisms that recruit existing biophysical features into higher-level architectures.
Conflict of interest statement
Figures

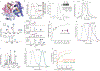
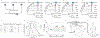


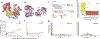








Comment in
-
Extinct proteins resurrected to reconstruct the evolution of vertebrate haemoglobin.Nature. 2020 May;581(7809):388-389. doi: 10.1038/d41586-020-01287-8. Nature. 2020. PMID: 32433630 No abstract available.
References
-
- Ahnert SE, Marsh JA, Hernández H, Robinson CV & Teichmann SA Principles of assembly reveal a periodic table of protein complexes. Science (80-. ) 350, (2015). - PubMed
-
- Marsh JA & Teichmann SA Structure, Dynamics, Assembly, and Evolution of Protein Complexes. Annu. Rev. Biochem 84, 551–575 (2015). - PubMed
-
- Monod J, Wyman J, & Changeux JP On the nature of allosteric transitions: a plausible model. Jounal Mol. Biol 12, 88–118 (1965). - PubMed
-
- Goodsell DS & Olson AJ Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct 29, 105–153 (2000). - PubMed
REFERENCES FOR METHODS AND EXTENDED DATA
-
- Guindon S et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321 (2010). - PubMed
-
- Le SQ & Gascuel O An improved general amino acid replacement matrix. Mol. Biol. Evol 25, 1307–1320 (2008). - PubMed
-
- Anisimova M & Gascuel O Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol 55, 539–552 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

