Design of a Multiepitope-Based Peptide Vaccine against the E Protein of Human COVID-19: An Immunoinformatics Approach
- PMID: 32461973
- PMCID: PMC7212276
- DOI: 10.1155/2020/2683286
Design of a Multiepitope-Based Peptide Vaccine against the E Protein of Human COVID-19: An Immunoinformatics Approach
Abstract
Background: A new endemic disease has spread across Wuhan City, China, in December 2019. Within few weeks, the World Health Organization (WHO) announced a novel coronavirus designated as coronavirus disease 2019 (COVID-19). In late January 2020, WHO declared the outbreak of a "public-health emergency of international concern" due to the rapid and increasing spread of the disease worldwide. Currently, there is no vaccine or approved treatment for this emerging infection; thus, the objective of this study is to design a multiepitope peptide vaccine against COVID-19 using an immunoinformatics approach.
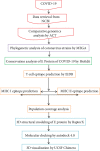
Method: Several techniques facilitating the combination of the immunoinformatics approach and comparative genomic approach were used in order to determine the potential peptides for designing the T-cell epitope-based peptide vaccine using the envelope protein of 2019-nCoV as a target.
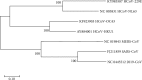
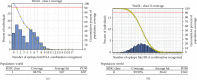
Results: Extensive mutations, insertion, and deletion were discovered with comparative sequencing in the COVID-19 strain. Additionally, ten peptides binding to MHC class I and MHC class II were found to be promising candidates for vaccine design with adequate world population coverage of 88.5% and 99.99%, respectively.
Conclusion: The T-cell epitope-based peptide vaccine was designed for COVID-19 using the envelope protein as an immunogenic target. Nevertheless, the proposed vaccine rapidly needs to be validated clinically in order to ensure its safety and immunogenic profile to help stop this epidemic before it leads to devastating global outbreaks.
Copyright © 2020 Miyssa I. Abdelmageed et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Hui D. S., I Azhar E., Madani T. A., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health — The latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. - DOI - PMC - PubMed
-
- CoV2020. GISAID EpifluDB. Archived from the original on 12 January 2020. 2020.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

