Programmed genome rearrangements in ciliates
- PMID: 32462406
- PMCID: PMC7599177
- DOI: 10.1007/s00018-020-03555-2
Programmed genome rearrangements in ciliates
Abstract
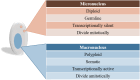
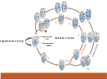
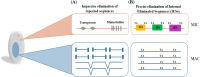
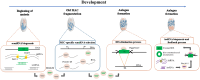
Ciliates are a highly divergent group of unicellular eukaryotes with separate somatic and germline genomes found in distinct dimorphic nuclei. This characteristic feature is tightly linked to extremely laborious developmentally regulated genome rearrangements in the development of a new somatic genome/nuclei following sex. The transformation from germline to soma genome involves massive DNA elimination mediated by non-coding RNAs, chromosome fragmentation, as well as DNA amplification. In this review, we discuss the similarities and differences in the genome reorganization processes of the model ciliates Paramecium and Tetrahymena (class Oligohymenophorea), and the distantly related Euplotes, Stylonychia, and Oxytricha (class Spirotrichea).
Keywords: Ciliates; DNA elimination; Genome rearrangement; Nuclear dimorphism; Small non-coding RNAs.
Figures
References
-
- Boveri T. Ueber Differenzierung der Zellkerne wahrend der Furchung des Eies von Ascaris megalocephala. The first discovery and description of a DNA elimination, chromatin diminution, in the parasitic nematode Parascaris. Anat Anz. 1887;2:688–693.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

