A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem)
- PMID: 32462988
- PMCID: PMC7311162
- DOI: 10.1080/07391102.2020.1774419
A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem)
Abstract
The rapid global spread of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has created an unprecedented healthcare crisis. The treatment for the severe respiratory illness caused by this virus is primarily symptomatic at this point, although the usage of a broad antiviral drug Remdesivir has been allowed on emergency basis by the Food and Drug Administration (FDA). The ever-increasing death toll highlights an urgent need for development of specific antivirals. In this work, we have utilized docking and simulation methods to identify small molecule inhibitors of SARS-CoV-2 Membrane (M) and Envelope (E) proteins, which are essential for virus assembly and budding. A total of 70 compounds from an Indian medicinal plant source (Azadirachta indica or Neem) were virtually screened against these two proteins and further analyzed with molecular dynamics simulations, which resulted in the identification of a few common compounds with strong binding to both structural proteins. The compounds bind to biologically critical regions of M and E, indicating their potential to inhibit the functionality of these components. We hope that our computational approach may result in the identification of effective inhibitors of SARS-CoV-2 assembly.Communicated by Ramaswamy H. Sarma.
Keywords: MM-PBSA; SARS-CoV-2; docking; molecular dynamics simulation; natural compound.
Figures
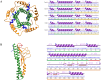

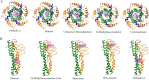

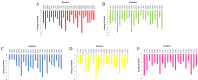
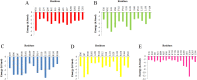
References
-
- Abdelli I., Hassani F., Bekkel Brikci S., & Ghalem S. (2020). In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1763199 - DOI - PMC - PubMed
-
- Akram M., Tahir I. M., Shah S. M. A., Mahmood Z., Altaf A., Ahmad K., Munir N., Daniyal M., Nasir S., & Mehboob H. (2018). Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: A systematic review. Phytotherapy Research: PTR, 32(5), 811–822. 10.1002/ptr.6024 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
