MEIOK21: a new component of meiotic recombination bridges required for spermatogenesis
- PMID: 32463460
- PMCID: PMC7337969
- DOI: 10.1093/nar/gkaa406
MEIOK21: a new component of meiotic recombination bridges required for spermatogenesis
Abstract
Repair of DNA double-strand breaks (DSBs) with homologous chromosomes is a hallmark of meiosis that is mediated by recombination 'bridges' between homolog axes. This process requires cooperation of DMC1 and RAD51 to promote homology search and strand exchange. The mechanism(s) regulating DMC1/RAD51-ssDNA nucleoprotein filament and the components of 'bridges' remain to be investigated. Here we show that MEIOK21 is a newly identified component of meiotic recombination bridges and is required for efficient formation of DMC1/RAD51 foci. MEIOK21 dynamically localizes on chromosomes from on-axis foci to 'hanging foci', then to 'bridges', and finally to 'fused foci' between homolog axes. Its chromosome localization depends on DSBs. Knockout of Meiok21 decreases the numbers of HSF2BP and DMC1/RAD51 foci, disrupting DSB repair, synapsis and crossover recombination and finally causing male infertility. Therefore, MEIOK21 is a novel recombination factor and probably mediates DMC1/RAD51 recruitment to ssDNA or their stability on chromosomes through physical interaction with HSF2BP.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
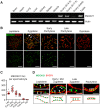
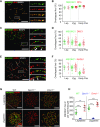


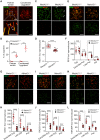
Similar articles
-
Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation.PLoS Genet. 2013;9(12):e1003978. doi: 10.1371/journal.pgen.1003978. Epub 2013 Dec 19. PLoS Genet. 2013. PMID: 24367271 Free PMC article.
-
Meiosis-Specific C19orf57/4930432K21Rik/BRME1 Modulates Localization of RAD51 and DMC1 to DSBs in Mouse Meiotic Recombination.Cell Rep. 2020 May 26;31(8):107686. doi: 10.1016/j.celrep.2020.107686. Cell Rep. 2020. PMID: 32460033
-
FIGNL1-FIRRM is essential for meiotic recombination and prevents DNA damage-independent RAD51 and DMC1 loading.Nat Commun. 2024 Aug 15;15(1):7015. doi: 10.1038/s41467-024-51458-8. Nat Commun. 2024. PMID: 39147779 Free PMC article.
-
Repair of DNA double-strand breaks in plant meiosis: role of eukaryotic RecA recombinases and their modulators.Plant Reprod. 2023 Mar;36(1):17-41. doi: 10.1007/s00497-022-00443-6. Epub 2022 Jun 1. Plant Reprod. 2023. PMID: 35641832 Review.
-
Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination.Cytogenet Genome Res. 2004;107(3-4):201-7. doi: 10.1159/000080598. Cytogenet Genome Res. 2004. PMID: 15467365 Review.
Cited by
-
A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1.Elife. 2020 Aug 26;9:e56996. doi: 10.7554/eLife.56996. Elife. 2020. PMID: 32845237 Free PMC article.
-
High Resolution View on the Regulation of Recombinase Accumulation in Mammalian Meiosis.Front Cell Dev Biol. 2021 May 24;9:672191. doi: 10.3389/fcell.2021.672191. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34109178 Free PMC article. Review.
-
Overexpression of HSF2 binding protein suppresses endoplasmic reticulum stress via regulating subcellular localization of CDC73 in hepatocytes.Cell Biosci. 2023 Mar 24;13(1):64. doi: 10.1186/s13578-023-01010-w. Cell Biosci. 2023. PMID: 36964632 Free PMC article.
-
ZFP541 and KCTD19 regulate chromatin organization and transcription programs for male meiotic progression.Cell Prolif. 2024 Apr;57(4):e13567. doi: 10.1111/cpr.13567. Epub 2023 Nov 3. Cell Prolif. 2024. PMID: 37921559 Free PMC article.
-
Testis-Specific PDHA2 Is Required for Proper Meiotic Recombination and Chromosome Organisation During Spermatogenesis.Cell Prolif. 2025 Jul;58(7):e70003. doi: 10.1111/cpr.70003. Epub 2025 Feb 20. Cell Prolif. 2025. PMID: 39973374 Free PMC article.
References
-
- Capalbo A., Hoffmann E.R., Cimadomo D., Ubaldi F.M., Rienzi L.. Human female meiosis revised: new insights into the mechanisms of chromosome segregation and aneuploidies from advanced genomics and time-lapse imaging. Hum. Reprod. Update. 2017; 23:706–722. - PubMed
-
- Keeney S., Giroux C.N., Kleckner N.. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997; 88:375–384. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

