Freedom of assembly: metabolic enzymes come together
- PMID: 32463766
- PMCID: PMC7353150
- DOI: 10.1091/mbc.E18-10-0675
Freedom of assembly: metabolic enzymes come together
Abstract
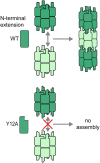
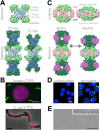
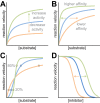
Many different enzymes in intermediate metabolism dynamically assemble filamentous polymers in cells, often in response to changes in physiological conditions. Most of the enzyme filaments known to date have only been observed in cells, but in a handful of cases structural and biochemical studies have revealed the mechanisms and consequences of assembly. In general, enzyme polymerization functions as a mechanism to allosterically tune enzyme kinetics, and it may play a physiological role in integrating metabolic signaling. Here, we highlight some principles of metabolic filaments by focusing on two well-studied examples in nucleotide biosynthesis pathways-inosine-5'-monophosphate (IMP) dehydrogenase and cytosine triphosphate (CTP) synthase.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

