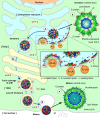
Capsid protein is central to the birth of flavivirus particles
- PMID: 32463839
- PMCID: PMC7255599
- DOI: 10.1371/journal.ppat.1008542
Capsid protein is central to the birth of flavivirus particles
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

