Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality?
- PMID: 32464271
- PMCID: PMC7247990
- DOI: 10.1016/j.ijid.2020.05.071
Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality?
Abstract
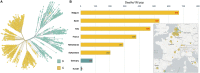
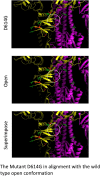
The increasing number of deaths due to the COVID-19 pandemic has raised serious global concerns. Increased testing capacity and ample intensive care availability could explain lower mortality in some countries compared to others. Nevertheless, it is also plausible that the SARS-CoV-2 mutations giving rise to different phylogenetic clades are responsible for the apparent death rate disparities around the world. Current research literature linking the genetic make-up of SARS-CoV-2 with fatalities is lacking. Here, we suggest that this disparity in fatality rates may be attributed to SARS-CoV-2 evolving mutations and urge the international community to begin addressing the phylogenetic clade classification of SARS-CoV-2 in relation to clinical outcomes.
Keywords: COVID-19; Coronavirus; D614G; S-Protein; SARS-CoV-2.
Copyright © 2020 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Next strain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. Available from: https://nextstrain.org/ncov. [Accessed 2 May 2020] - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

