Epigenetic Modifications: An Unexplored Facet of Exogenous RNA Application in Plants
- PMID: 32466487
- PMCID: PMC7356522
- DOI: 10.3390/plants9060673
Epigenetic Modifications: An Unexplored Facet of Exogenous RNA Application in Plants
Abstract
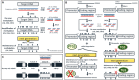
Exogenous RNA interference (exo-RNAi) is a powerful transgene-free tool in modern crop improvement and protection platforms. In exo-RNAi approaches, double-stranded RNAs (dsRNAs) or short-interfering RNAs (siRNAs) are externally applied in plants in order to selectively trigger degradation of target mRNAs. Yet, the applied dsRNAs may also trigger unintended epigenetic alterations and result in epigenetically modified plants, an issue that has not been sufficiently addressed and which merits more careful consideration.
Keywords: PTGS; RdDM; dsRNAs; epigenetics; exogenous RNAi; siRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The Specificity of Transgene Suppression in Plants by Exogenous dsRNA.Plants (Basel). 2022 Mar 8;11(6):715. doi: 10.3390/plants11060715. Plants (Basel). 2022. PMID: 35336598 Free PMC article.
-
Application of Exogenous dsRNAs-induced RNAi in Agriculture: Challenges and Triumphs.Front Plant Sci. 2020 Jun 25;11:946. doi: 10.3389/fpls.2020.00946. eCollection 2020. Front Plant Sci. 2020. PMID: 32670336 Free PMC article. Review.
-
RNA interference: the molecular immune system.J Mol Histol. 2004 Aug;35(6):545-53. doi: 10.1007/s10735-004-2192-8. J Mol Histol. 2004. PMID: 15614608 Review.
-
Classification of and detection techniques for RNAi-induced effects in GM plants.Front Plant Sci. 2025 Mar 7;16:1535384. doi: 10.3389/fpls.2025.1535384. eCollection 2025. Front Plant Sci. 2025. PMID: 40123947 Free PMC article. Review.
-
Analysis of short interfering RNA function in RNA interference by using Drosophila embryo extracts and schneider cells.Methods Enzymol. 2005;392:351-71. doi: 10.1016/S0076-6879(04)92021-6. Methods Enzymol. 2005. PMID: 15644192
Cited by
-
Exogenous Application of dsRNA-Inducing Silencing of the Fusarium oxysporum Tup1 Gene and Reducing Its Virulence.Int J Mol Sci. 2024 Sep 24;25(19):10286. doi: 10.3390/ijms251910286. Int J Mol Sci. 2024. PMID: 39408614 Free PMC article.
-
Applications of Virus-Induced Gene Silencing in Cotton.Plants (Basel). 2024 Jan 17;13(2):272. doi: 10.3390/plants13020272. Plants (Basel). 2024. PMID: 38256825 Free PMC article. Review.
-
Concepts and considerations for enhancing RNAi efficiency in phytopathogenic fungi for RNAi-based crop protection using nanocarrier-mediated dsRNA delivery systems.Front Fungal Biol. 2022 Sep 8;3:977502. doi: 10.3389/ffunb.2022.977502. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746174 Free PMC article. Review.
-
Lab-to-Field Transition of RNA Spray Applications - How Far Are We?Front Plant Sci. 2021 Oct 15;12:755203. doi: 10.3389/fpls.2021.755203. eCollection 2021. Front Plant Sci. 2021. PMID: 34721485 Free PMC article. Review.
-
Exploiting Epigenetic Variations for Crop Disease Resistance Improvement.Front Plant Sci. 2021 Jun 4;12:692328. doi: 10.3389/fpls.2021.692328. eCollection 2021. Front Plant Sci. 2021. PMID: 34149790 Free PMC article. Review.
References
-
- Fusaro A.F., Matthew L., Smith N.A., Curtin S.J., Dedic-Hagan J., Ellacott G.A., Watson J.M., Wang M.B., Brosnan C., Carroll B.J., et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

