Comparing the Influence of Assembly Processes Governing Bacterial Community Succession Based on DNA and RNA Data
- PMID: 32466517
- PMCID: PMC7355735
- DOI: 10.3390/microorganisms8060798
Comparing the Influence of Assembly Processes Governing Bacterial Community Succession Based on DNA and RNA Data
Abstract
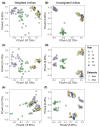
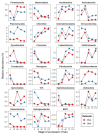
Quantifying which assembly processes structure microbiomes can assist prediction, manipulation, and engineering of community outcomes. However, the relative importance of these processes might depend on whether DNA or RNA are used, as they differ in stability. We hypothesized that. RNA-inferred community responses to (a)biotic fluctuations are faster than those inferred by DNA; the relative influence of variable selection is stronger in RNA-inferred communities (environmental factors are spatiotemporally heterogeneous), whereas homogeneous selection largely influences DNA-inferred communities (environmental filters are constant). To test these hypotheses, we characterized soil bacterial communities by sequencing both 16S rRNA amplicons from the extracted DNA and RNA transcripts across distinct stages of soil primary succession and quantified the relative influence of each assembly process using ecological null model analysis. Our results revealed that variations in α-diversity and temporal turnover were higher in RNA- than in DNA-inferred communities across successional stages, albeit there was a similar community composition; in line with our hypotheses, the assembly of RNA-inferred community was more closely associated with environmental variability (variable selection) than using the standard DNA-based approach, which was largely influenced by homogeneous selection. This study illustrates the need for benchmarking approaches to properly elucidate how community assembly processes structure microbial communities.
Keywords: 16S rRNA gene; community turnover; dispersal; drift; ecological modeling; selection.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Castle S.C., Nemergut D.R., Grandy A.S., Leff J.W., Graham E.B., Hood E., Schmidt S.K., Wickings K., Cleveland C.C. Biogeochemical drivers of microbial community convergence across actively retreating glaciers. Soil Biol. Biochem. 2016;101:74–84. doi: 10.1016/j.soilbio.2016.07.010. - DOI
LinkOut - more resources
Full Text Sources

