Canonical Notch signaling controls the early thymic epithelial progenitor cell state and emergence of the medullary epithelial lineage in fetal thymus development
- PMID: 32467237
- PMCID: PMC7328009
- DOI: 10.1242/dev.178582
Canonical Notch signaling controls the early thymic epithelial progenitor cell state and emergence of the medullary epithelial lineage in fetal thymus development
Abstract
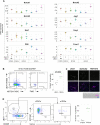
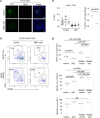
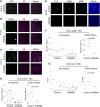
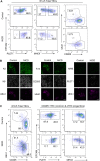
Thymus function depends on the epithelial compartment of the thymic stroma. Cortical thymic epithelial cells (cTECs) regulate T cell lineage commitment and positive selection, while medullary (m) TECs impose central tolerance on the T cell repertoire. During thymus organogenesis, these functionally distinct sub-lineages are thought to arise from a common thymic epithelial progenitor cell (TEPC). However, the mechanisms controlling cTEC and mTEC production from the common TEPC are not understood. Here, we show that emergence of the earliest mTEC lineage-restricted progenitors requires active NOTCH signaling in progenitor TEC and that, once specified, further mTEC development is NOTCH independent. In addition, we demonstrate that persistent NOTCH activity favors maintenance of undifferentiated TEPCs at the expense of cTEC differentiation. Finally, we uncover a cross-regulatory relationship between NOTCH and FOXN1, a master regulator of TEC differentiation. These data establish NOTCH as a potent regulator of TEPC and mTEC fate during fetal thymus development, and are thus of high relevance to strategies aimed at generating/regenerating functional thymic tissue in vitro and in vivo.
Keywords: Cell fate regulation; Differentiation; Lineage divergence; Notch signaling; Progenitor cell; Stem cell; Thymic epithelial cell; Thymus.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


Similar articles
-
NOTCH1 signaling establishes the medullary thymic epithelial cell progenitor pool during mouse fetal development.Development. 2020 Jun 22;147(12):dev178988. doi: 10.1242/dev.178988. Development. 2020. PMID: 32467240 Free PMC article.
-
Long-term persistence of functional thymic epithelial progenitor cells in vivo under conditions of low FOXN1 expression.PLoS One. 2014 Dec 22;9(12):e114842. doi: 10.1371/journal.pone.0114842. eCollection 2014. PLoS One. 2014. PMID: 25531271 Free PMC article.
-
Abrogation of Notch Signaling in Embryonic TECs Impacts Postnatal mTEC Homeostasis and Thymic Involution.Front Immunol. 2022 May 30;13:867302. doi: 10.3389/fimmu.2022.867302. eCollection 2022. Front Immunol. 2022. PMID: 35707539 Free PMC article.
-
Serial progression of cortical and medullary thymic epithelial microenvironments.Eur J Immunol. 2014 Jan;44(1):16-22. doi: 10.1002/eji.201344110. Epub 2013 Dec 4. Eur J Immunol. 2014. PMID: 24214487 Free PMC article. Review.
-
Adult thymic epithelial cell (TEC) progenitors and TEC stem cells: Models and mechanisms for TEC development and maintenance.Eur J Immunol. 2015 Nov;45(11):2985-93. doi: 10.1002/eji.201545844. Epub 2015 Sep 30. Eur J Immunol. 2015. PMID: 26362014 Review.
Cited by
-
Generation and repair of thymic epithelial cells.J Exp Med. 2024 Oct 7;221(10):e20230894. doi: 10.1084/jem.20230894. Epub 2024 Jul 9. J Exp Med. 2024. PMID: 38980292 Free PMC article. Review.
-
Mechanisms underlying the direct programming of mouse embryonic fibroblasts to thymic epithelial cells by FOXN1.Development. 2024 Jul 15;151(14):dev202730. doi: 10.1242/dev.202730. Epub 2024 Jul 22. Development. 2024. PMID: 38958026 Free PMC article.
-
Notch Partners in the Long Journey of T-ALL Pathogenesis.Int J Mol Sci. 2023 Jan 10;24(2):1383. doi: 10.3390/ijms24021383. Int J Mol Sci. 2023. PMID: 36674902 Free PMC article. Review.
-
Phylogeny, Structure, Functions, and Role of AIRE in the Formation of T-Cell Subsets.Cells. 2022 Jan 7;11(2):194. doi: 10.3390/cells11020194. Cells. 2022. PMID: 35053310 Free PMC article. Review.
-
Developmental dynamics of two bipotent thymic epithelial progenitor types.Nature. 2022 Jun;606(7912):165-171. doi: 10.1038/s41586-022-04752-8. Epub 2022 May 25. Nature. 2022. PMID: 35614226 Free PMC article.
References
-
- Akiyama N., Takizawa N., Miyauchi M., Yanai H., Tateishi R., Shinzawa M., Yoshinaga R., Kurihara M., Demizu Y., Yasuda H. et al. (2016). Identification of embryonic precursor cells that differentiate into thymic epithelial cells expressing autoimmune regulator. J. Exp. Med. 213, 1441-1458. 10.1084/jem.20151780 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

