A combinatorially regulated RNA splicing signature predicts breast cancer EMT states and patient survival
- PMID: 32467311
- PMCID: PMC7430667
- DOI: 10.1261/rna.074187.119
A combinatorially regulated RNA splicing signature predicts breast cancer EMT states and patient survival
Abstract
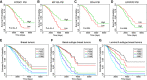
During breast cancer metastasis, the developmental process epithelial-mesenchymal transition (EMT) is abnormally activated. Transcriptional regulatory networks controlling EMT are well-studied; however, alternative RNA splicing also plays a critical regulatory role during this process. A comprehensive understanding of alternative splicing (AS) and the RNA binding proteins (RBPs) that regulate it during EMT and their impact on breast cancer remains largely unknown. In this study, we annotated AS in the breast cancer TCGA data set and identified an AS signature that is capable of distinguishing epithelial and mesenchymal states of the tumors. This AS signature contains 25 AS events, among which nine showed increased exon inclusion and 16 showed exon skipping during EMT. This AS signature accurately assigns the EMT status of cells in the CCLE data set and robustly predicts patient survival. We further developed an effective computational method using bipartite networks to identify RBP-AS networks during EMT. This network analysis revealed the complexity of RBP regulation and nominated previously unknown RBPs that regulate EMT-associated AS events. This study highlights the importance of global AS regulation during EMT in cancer progression and paves the way for further investigation into RNA regulation in EMT and metastasis.
Keywords: RNA-binding proteins (RBPs); alternative splicing (AS); breast cancer; epithelial–mesenchymal transition (EMT).
© 2020 Qiu et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




References
-
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech 2008: P10008 10.1088/1742-5468/2008/10/P10008 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
