Transcriptome-Wide Comparisons and Virulence Gene Polymorphisms of Host-Associated Genotypes of the Cnidarian Parasite Ceratonova shasta in Salmonids
- PMID: 32467979
- PMCID: PMC7487138
- DOI: 10.1093/gbe/evaa109
Transcriptome-Wide Comparisons and Virulence Gene Polymorphisms of Host-Associated Genotypes of the Cnidarian Parasite Ceratonova shasta in Salmonids
Abstract
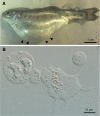
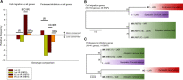
Ceratonova shasta is an important myxozoan pathogen affecting the health of salmonid fishes in the Pacific Northwest of North America. Ceratonova shasta exists as a complex of host-specific genotypes, some with low to moderate virulence, and one that causes a profound, lethal infection in susceptible hosts. High throughput sequencing methods are powerful tools for discovering the genetic basis of these host/virulence differences, but deep sequencing of myxozoans has been challenging due to extremely fast molecular evolution of this group, yielding strongly divergent sequences that are difficult to identify, and unavoidable host contamination. We designed and optimized different bioinformatic pipelines to address these challenges. We obtained a unique set of comprehensive, host-free myxozoan RNA-seq data from C. shasta genotypes of varying virulence from different salmonid hosts. Analyses of transcriptome-wide genetic distances and maximum likelihood multigene phylogenies elucidated the evolutionary relationship between lineages and demonstrated the limited resolution of the established Internal Transcribed Spacer marker for C. shasta genotype identification, as this marker fails to differentiate between biologically distinct genotype II lineages from coho salmon and rainbow trout. We further analyzed the data sets based on polymorphisms in two gene groups related to virulence: cell migration and proteolytic enzymes including their inhibitors. The developed single-nucleotide polymorphism-calling pipeline identified polymorphisms between genotypes and demonstrated that variations in both motility and protease genes were associated with different levels of virulence of C. shasta in its salmonid hosts. The prospective use of proteolytic enzymes as promising candidates for targeted interventions against myxozoans in aquaculture is discussed. We developed host-free transcriptomes of a myxozoan model organism from strains that exhibited different degrees of virulence, as a unique source of data that will foster functional gene analyses and serve as a base for the development of potential therapeutics for efficient control of these parasites.
Keywords: Myxozoa; SNPs; aquaculture; cell migration/motility; proteases.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Atkinson SD, Bartholomew JL.. 2010. a. Disparate infection patterns of Ceratomyxa shasta (Myxozoa) in rainbow trout Oncorhynchus mykiss and Chinook salmon Oncorhynchus tshawytscha correlate with Internal Transcribed Spacer-1 sequence variation in the parasite. Int J Parasitol. 40(5):599–604. - PubMed
-
- Atkinson SD, Bartholomew JL.. 2010. b. Spatial, temporal and host factors structure the Ceratomyxa shasta (Myxozoa) population in the Klamath River basin. Infect Genet Evol. 10(7):1019–1026. - PubMed
-
- Atkinson SD, Bartholomew JL, Lotan T.. 2018. Myxozoans: ancient metazoan parasites find a home in phylum Cnidaria. Zoology 129:66–68. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

