Pan‑cancer analysis of transmembrane protease serine 2 and cathepsin L that mediate cellular SARS‑CoV‑2 infection leading to COVID-19
- PMID: 32468052
- PMCID: PMC7307597
- DOI: 10.3892/ijo.2020.5071
Pan‑cancer analysis of transmembrane protease serine 2 and cathepsin L that mediate cellular SARS‑CoV‑2 infection leading to COVID-19
Abstract
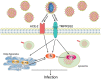
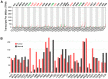
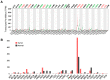
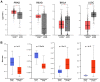
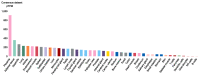
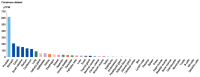
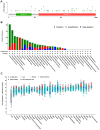
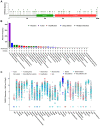
Severe acute respiratory syndrome (SARS) coronavirus‑2 (SARS‑CoV2) is the cause of a new disease (COVID‑19) which has evolved into a pandemic during the first half of 2020. Older age, male sex and certain underlying diseases, including cancer, appear to significantly increase the risk for severe COVID‑19. SARS‑CoV‑2 infection of host cells is facilitated by the angiotensin‑converting enzyme 2 (ACE‑2), and by transmembrane protease serine 2 (TMPRSS2) and other host cell proteases such as cathepsin L (CTSL). With the exception of ACE‑2, a systematic analysis of these two other SARS‑CoV2 infection mediators in malignancies is lacking. Here, we analysed genetic alteration, RNA expression, and DNA methylation of TMPRSS2 and CTSL across a wide spectrum of tumors and controls. TMPRSS2 was overexpressed in cervical squamous cell carcinoma and endocervical adenocarcinoma, colon adenocarcinoma, prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), uterine corpus endometrial carcinoma and uterine carcinosarcoma, with PRAD and READ exhibiting the highest expression of all cancers. CTSL was upregulated in lymphoid neoplasm diffuse large B‑cell lymphoma, oesophageal carcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, lower grade glioma, pancreatic adenocarcinoma, skin cutaneous melanoma, stomach adenocarcinoma, and thymoma. Hypo‑methylation of both genes was evident in most cases where they have been highly upregulated. We have expanded on our observations by including data relating to mutations and copy number alterations at pan‑cancer level. The novel hypotheses that are stemming out of these data need to be further investigated and validated in large clinical studies.
Keywords: cathepsin L (CTSL); transmembrane protease serine 2 (TMPRSS2); pan-cancer; DNA methylation; COVID-19; angiotensin-converting enzyme 2.
Figures

References
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
