Integrated Profiling Revealed Potential Regulatory Networks Among Long Noncoding RNAs and mRNAs in Mucosal Gastric Cancer
- PMID: 32468960
- PMCID: PMC7268121
- DOI: 10.1177/1533033820930119
Integrated Profiling Revealed Potential Regulatory Networks Among Long Noncoding RNAs and mRNAs in Mucosal Gastric Cancer
Abstract
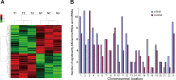
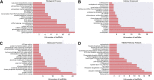
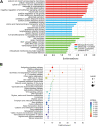
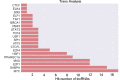
Gastric cancer is one of the most commonly occurring cancers worldwide. Investigation of long noncoding RNAs is of increasing interest, particularly in relation to their contribution to progression and prognosis of gastric cancers; however, insufficient studies been performed investigating the part of long noncoding RNAs play in gastric cancer carcinogenesis. Patterns of dysregulated long noncoding RNA and messenger RNA between mucosa gastric cancer and adjacent normal tissues were identified using long noncoding RNAs microarray analysis. Quantitative real-time polymerase chain reaction was conducted as a means to verify the obtained data. Both Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were subsequently used to investigate the function of dysregulated long noncoding RNAs and messenger RNAs. Cis and trans action was used to predict the possible targets of long noncoding RNAs, and a coexpression network was created to simulate the complex intergenic interactions. Ninety-five dysregulated long noncoding RNAs and 123 messenger RNAs were identified, and quantitative real-time polymerase chain reaction was used to validate 6 filtered long noncoding RNAs. Gene Ontology and KEGG pathway analyses identified several remarkably biological processes and signaling pathways, including spliceosome, RNA transport, and ubiquitin-mediated proteolysis. The transcriptional factors MYC, GABPA, and E2F1 were found to play a central function in the long noncoding RNAs process, as indicated by the coexpression network. This study revealed the dysregulated long noncoding RNA profiles of mucosal gastric cancer. The results shed light on the biological function of long noncoding RNAs in gastric cancer pathogenesis. This provides useful information for exploring potential early screening biomarkers in gastric cancer.
Keywords: expression profiles; long noncoding RNA (lncRNA); mucosal gastric cancer.
Conflict of interest statement
Figures


Similar articles
-
Comprehensive analysis of expression profiles of long non‑coding RNAs with associated ceRNA network involved in gastric cancer progression.Mol Med Rep. 2019 Sep;20(3):2209-2218. doi: 10.3892/mmr.2019.10478. Epub 2019 Jul 9. Mol Med Rep. 2019. PMID: 31322220 Free PMC article.
-
Genome-Wide Analysis Identified a Number of Dysregulated Long Noncoding RNA (lncRNA) in Human Pancreatic Ductal Adenocarcinoma.Technol Cancer Res Treat. 2018 Jan 1;17:1533034617748429. doi: 10.1177/1533034617748429. Technol Cancer Res Treat. 2018. PMID: 29343207 Free PMC article.
-
Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma.Oncotarget. 2017 Jul 25;8(30):49201-49216. doi: 10.18632/oncotarget.17329. Oncotarget. 2017. PMID: 28484081 Free PMC article.
-
Long noncoding RNAs in gastric cancer carcinogenesis and metastasis.Brief Funct Genomics. 2017 May 1;16(3):129-145. doi: 10.1093/bfgp/elw011. Brief Funct Genomics. 2017. PMID: 27122631 Review.
-
Role of circular RNAs and long non‑coding RNAs in the clinical translation of gastric cancer (Review).Int J Mol Med. 2021 Jan;47(1):77-91. doi: 10.3892/ijmm.2020.4774. Epub 2020 Oct 29. Int J Mol Med. 2021. PMID: 33416088 Free PMC article. Review.
Cited by
-
Identification of lncRNA and mRNA regulatory networks associated with gastric cancer progression.Front Oncol. 2023 Mar 6;13:1140460. doi: 10.3389/fonc.2023.1140460. eCollection 2023. Front Oncol. 2023. PMID: 36969001 Free PMC article.
References
-
- Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388(10060):2654–2664. - PubMed
-
- Coburn N, Cosby R, Klein L, et al. Staging and surgical approaches in gastric cancer: a systematic review. Cancer Treat Rev. 2018;63:104–115. - PubMed
-
- Moehler M, Baltin CT, Ebert M, et al. International comparison of the German evidence-based S3-guidelines on the diagnosis and multimodal treatment of early and locally advanced gastric cancer, including adenocarcinoma of the lower esophagus. Gastric Cancer. 2015;18(3):550–563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

