mCSM-membrane: predicting the effects of mutations on transmembrane proteins
- PMID: 32469063
- PMCID: PMC7319563
- DOI: 10.1093/nar/gkaa416
mCSM-membrane: predicting the effects of mutations on transmembrane proteins
Abstract
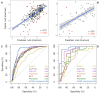
Significant efforts have been invested into understanding and predicting the molecular consequences of mutations in protein coding regions, however nearly all approaches have been developed using globular, soluble proteins. These methods have been shown to poorly translate to studying the effects of mutations in membrane proteins. To fill this gap, here we report, mCSM-membrane, a user-friendly web server that can be used to analyse the impacts of mutations on membrane protein stability and the likelihood of them being disease associated. mCSM-membrane derives from our well-established mutation modelling approach that uses graph-based signatures to model protein geometry and physicochemical properties for supervised learning. Our stability predictor achieved correlations of up to 0.72 and 0.67 (on cross validation and blind tests, respectively), while our pathogenicity predictor achieved a Matthew's Correlation Coefficient (MCC) of up to 0.77 and 0.73, outperforming previously described methods in both predicting changes in stability and in identifying pathogenic variants. mCSM-membrane will be an invaluable and dedicated resource for investigating the effects of single-point mutations on membrane proteins through a freely available, user friendly web server at http://biosig.unimelb.edu.au/mcsm_membrane.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Overington J.P., Al-Lazikani B., Hopkins A.L.. How many drug targets are there. Nat. Rev. Drug Discov. 2006; 5:993–996. - PubMed
-
- Frishman D., Mewes H.W.. Protein structural classes in five complete genomes. Nat. Struct. Biol. 1997; 4:626–628. - PubMed
-
- Fagerberg L., Jonasson K., von Heijne G., Uhlen M., Berglund L.. Prediction of the human membrane proteome. Proteomics. 2010; 10:1141–1149. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

