Spatial inter-centromeric interactions facilitated the emergence of evolutionary new centromeres
- PMID: 32469306
- PMCID: PMC7292649
- DOI: 10.7554/eLife.58556
Spatial inter-centromeric interactions facilitated the emergence of evolutionary new centromeres
Abstract
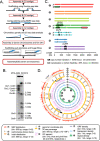
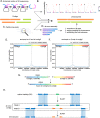
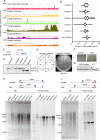
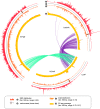
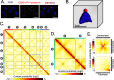
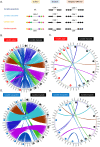
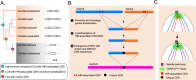
Centromeres of Candida albicans form on unique and different DNA sequences but a closely related species, Candida tropicalis, possesses homogenized inverted repeat (HIR)-associated centromeres. To investigate the mechanism of centromere type transition, we improved the fragmented genome assembly and constructed a chromosome-level genome assembly of C. tropicalis by employing PacBio sequencing, chromosome conformation capture sequencing (3C-seq), chromoblot, and genetic analysis of engineered aneuploid strains. Further, we analyzed the 3D genome organization using 3C-seq data, which revealed spatial proximity among the centromeres as well as telomeres of seven chromosomes in C. tropicalis. Intriguingly, we observed evidence of inter-centromeric translocations in the common ancestor of C. albicans and C. tropicalis. Identification of putative centromeres in closely related Candida sojae, Candida viswanathii and Candida parapsilosis indicates loss of ancestral HIR-associated centromeres and establishment of evolutionary new centromeres (ENCs) in C. albicans. We propose that spatial proximity of the homologous centromere DNA sequences facilitated karyotype rearrangements and centromere type transitions in human pathogenic yeasts of the CUG-Ser1 clade.
Keywords: 3D-genome; CUG-Ser1 clade; Candida; chromosome segregation; chromosomes; evolutionary new centromere; gene expression; genetics; genomics; karyotype rearrangement.
© 2020, Guin et al.
Conflict of interest statement
KG, YC, RM, SM, BT, CO, GB, AS, KS No competing interests declared
Figures








Similar articles
-
Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis.Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19797-802. doi: 10.1073/pnas.0809770105. Epub 2008 Dec 5. Proc Natl Acad Sci U S A. 2008. PMID: 19060206 Free PMC article.
-
Repeat-Associated Fission Yeast-Like Regional Centromeres in the Ascomycetous Budding Yeast Candida tropicalis.PLoS Genet. 2016 Feb 4;12(2):e1005839. doi: 10.1371/journal.pgen.1005839. eCollection 2016 Feb. PLoS Genet. 2016. PMID: 26845548 Free PMC article.
-
Functional and Comparative Analysis of Centromeres Reveals Clade-Specific Genome Rearrangements in Candida auris and a Chromosome Number Change in Related Species.mBio. 2021 May 11;12(3):e00905-21. doi: 10.1128/mBio.00905-21. mBio. 2021. PMID: 33975937 Free PMC article.
-
Implications of the Evolutionary Trajectory of Centromeres in the Fungal Kingdom.Annu Rev Microbiol. 2020 Sep 8;74:835-853. doi: 10.1146/annurev-micro-011720-122512. Epub 2020 Jul 24. Annu Rev Microbiol. 2020. PMID: 32706633 Review.
-
Genetic and epigenetic effects on centromere establishment.Chromosoma. 2020 Mar;129(1):1-24. doi: 10.1007/s00412-019-00727-3. Epub 2019 Nov 28. Chromosoma. 2020. PMID: 31781852 Review.
Cited by
-
Interactions of Both Pathogenic and Nonpathogenic CUG Clade Candida Species with Macrophages Share a Conserved Transcriptional Landscape.mBio. 2021 Dec 21;12(6):e0331721. doi: 10.1128/mbio.03317-21. Epub 2021 Dec 14. mBio. 2021. PMID: 34903044 Free PMC article.
-
Candida albicans isolates contain frequent heterozygous structural variants and transposable elements within genes and centromeres.Genome Res. 2025 Apr 14;35(4):824-838. doi: 10.1101/gr.279301.124. Genome Res. 2025. PMID: 39438112
-
Stable Positions of Epigenetically Inherited Centromeres in the Emerging Fungal Pathogen Candida auris and Its Relatives.mBio. 2021 Aug 31;12(4):e0103621. doi: 10.1128/mBio.01036-21. Epub 2021 Jul 6. mBio. 2021. PMID: 34225489 Free PMC article.
-
Advances in understanding the evolution of fungal genome architecture.F1000Res. 2020 Jul 27;9:F1000 Faculty Rev-776. doi: 10.12688/f1000research.25424.1. eCollection 2020. F1000Res. 2020. PMID: 32765832 Free PMC article. Review.
-
Using Genomics to Shape the Definition of the Agglutinin-Like Sequence (ALS) Family in the Saccharomycetales.Front Cell Infect Microbiol. 2021 Dec 14;11:794529. doi: 10.3389/fcimb.2021.794529. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34970511 Free PMC article.
References
-
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA. Punctuated evolution of prostate Cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Shyama Prasad Mukherjee Fellowship 07/733(0181)/2013-EMR-I/Council of Scientific and Industrial Research/International
- BT/PR27490/Med/29/1323/2018/Department of Biotechnology , Ministry of Science and Technology/International
- RG39/18/Ministry of Education - Singapore/International
- Nanyang Assistant Professorship grant/Nanyang Technological University/International
- BT/PR27490/Med/29/1323/2018/Department of Biotechnology, Ministry of Science and Technology/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

