Variability in Cytogenetic Testing for Multiple Myeloma: A Comprehensive Analysis From Across the United States
- PMID: 32469686
- PMCID: PMC7564132
- DOI: 10.1200/JOP.19.00639
Variability in Cytogenetic Testing for Multiple Myeloma: A Comprehensive Analysis From Across the United States
Abstract
Purpose: Multiple myeloma (MM) treatment has changed tremendously, with significant improvement in patient out-comes. One group with a suboptimal benefit is patients with high-risk cytogenetics, as tested by conventional karyotyping or fluorescence in situ hybridization (FISH). Methodology for these tests has been published, but not necessarily standardized.
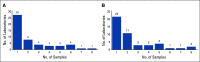
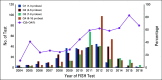
Methods: We address variability in the testing and reporting methodology for MM cytogenetics in the United States using the ongoing African American Multiple Myeloma Study (AAMMS). We evaluated clinical and cytogenetic data from 1,221 patients (1,161 with conventional karyotyping and 976 with FISH) tested between 1998 and 2016 across 58 laboratories nationwide.
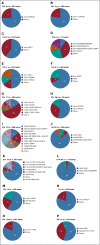
Results: Interlab and intralab variability was noted for the number of cells analyzed for karyotyping, with a significantly higher number of cells analyzed in patients in whom cytogenetics were normal (P 5.0025). For FISH testing, CD138-positive cell enrichment was used in 29.7% of patients and no enrichment in 50% of patients, whereas the remainder had unknown status. A significantly smaller number of cells was analyzed for patients in which CD138 cell enrichment was used compared with those without such enrichment (median, 50 v 200; P, .0001). A median of 7 loci probes (range, 1-16) were used for FISH testing across all laboratories, with variability in the loci probed even within a given laboratory. Chromosome 13-related abnormalities were the most frequently tested abnormality (n5956; 97.9%), and t(14;16) was the least frequently tested abnormality (n 5 119; 12.2%).
Conclusions: We report significant variability in cytogenetic testing across the United States for MM, potentially leading to variability in risk stratification, with possible clinical implications and personalized treatment approaches.
Conflict of interest statement
Variability in Cytogenetic Testing for Multiple Myeloma: A Comprehensive Analysis from Across the United States
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
Niquelle Brown Wade
Ajay K. Nooka
Daniel Stram
Graham A. Colditz
Jayesh Mehta
Howard Terebelo
Seema Singhal
Ravi Vij
Leon Bernal-Mizrachi
Jeffrey A. Zonder
Carol A. Huff
Sagar Lonial
Robert Z. Orlowski
Sikander Ailawadhi
No other potential conflicts of interest were reported.
Figures

References
-
- Siegel RL, Miller KD, Jemal A.Cancer statistics, 2018 CA Cancer J Clin 687–302018 - PubMed
-
- Ailawadhi S, Aldoss IT, Yang D, et al. Outcome disparities in multiple myeloma: A SEER-based comparative analysis of ethnic subgroups Br J Haematol 15891–982012 - PubMed
-
- Chan HSH, Chen CI, Reece DE.Current review on high-risk multiple myeloma Curr Hematol Malig Rep 1296–1082017 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

