Extensive Remodeling of the Immune Microenvironment in B Cell Acute Lymphoblastic Leukemia
- PMID: 32470390
- PMCID: PMC7341535
- DOI: 10.1016/j.ccell.2020.04.015
Extensive Remodeling of the Immune Microenvironment in B Cell Acute Lymphoblastic Leukemia
Abstract
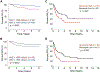
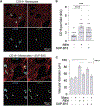
A subset of B cell acute lymphoblastic leukemia (B-ALL) patients will relapse and succumb to therapy-resistant disease. The bone marrow microenvironment may support B-ALL progression and treatment evasion. Utilizing single-cell approaches, we demonstrate B-ALL bone marrow immune microenvironment remodeling upon disease initiation and subsequent re-emergence during conventional chemotherapy. We uncover a role for non-classical monocytes in B-ALL survival, and demonstrate monocyte abundance at B-ALL diagnosis is predictive of pediatric and adult B-ALL patient survival. We show that human B-ALL blasts alter a vascularized microenvironment promoting monocytic differentiation, while depleting leukemia-associated monocytes in B-ALL animal models prolongs disease remission in vivo. Our profiling of the B-ALL immune microenvironment identifies extrinsic regulators of B-ALL survival supporting new immune-based therapeutic approaches for high-risk B-ALL treatment.
Keywords: acute lymphoblastic leukemia; chemotherapy; immune microenvironment; monocytes; relapse; single cell.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, and Iruela-Arispe ML (2006). VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn 235, 759–767. - PubMed
-
- Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, and Buettner F. (2016). destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

