MAX Functions as a Tumor Suppressor and Rewires Metabolism in Small Cell Lung Cancer
- PMID: 32470392
- PMCID: PMC7363581
- DOI: 10.1016/j.ccell.2020.04.016
MAX Functions as a Tumor Suppressor and Rewires Metabolism in Small Cell Lung Cancer
Abstract
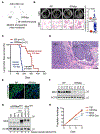
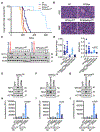
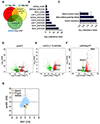
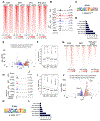
Small cell lung cancer (SCLC) is a highly aggressive and lethal neoplasm. To identify candidate tumor suppressors we applied CRISPR/Cas9 gene inactivation screens to a cellular model of early-stage SCLC. Among the top hits was MAX, the obligate heterodimerization partner for MYC family proteins that is mutated in human SCLC. Max deletion increases growth and transformation in cells and dramatically accelerates SCLC progression in an Rb1/Trp53-deleted mouse model. In contrast, deletion of Max abrogates tumorigenesis in MYCL-overexpressing SCLC. Max deletion in SCLC resulted in derepression of metabolic genes involved in serine and one-carbon metabolism. By increasing serine biosynthesis, Max-deleted cells exhibit resistance to serine depletion. Thus, Max loss results in metabolic rewiring and context-specific tumor suppression.
Keywords: CRISPR-Cas9 genetic screens; MAX; MYC; SCLC; Transcriptional regulation; cancer; mouse model; serine and one-carbon metabolism; small cell lung cancer; tumor suppressor genes.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests R.N.E. is a member of the Scientific Advisory Boards of Kronos Bio, Inc., and Shenogen Beijing.
Figures



References
-
- Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, and Land H (1993). Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72, 233–245. - PubMed
-
- Ayer DE, Lawrence QA, and Eisenman RN (1995). Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80, 767–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

