Identifying modifier genes for hypertrophic cardiomyopathy
- PMID: 32470469
- PMCID: PMC9768851
- DOI: 10.1016/j.yjmcc.2020.05.006
Identifying modifier genes for hypertrophic cardiomyopathy
Abstract
Background: Hypertrophic cardiomyopathy (HCM) severity greatly varies among patients even with the same HCM gene mutations. This variation is largely regulated by modifier gene(s), which, however, remain largely unknown. The current study is aimed to identify modifier genes using BXD strains, a large murine genetic reference population (GRP) derived from crosses between C57BL/6 J (B6) and D2 DBA/2 J (D2) mice. D2 mice natualy carrythe genetic basis and phenotypes of HCM.
Methods: Myocardial hypertrophy, the major phenotype of HCM, was determined by cardiomyocyte size on cardiac sections in 30 BXD strains, and their parental B6 and D2 strains and morphometric analysis was performed. Quantitative Trait Locus (QTL) mapping for cardiomyocyte sizes was conducted with WebQTL in GeneNetwork. Correlation of cardiomyocyte size and cardiac gene expression in BXDs accessed from GeneNetwork were evaluated. QTL candidate genes associated with cardiomyocyte sizes were prioritized based on the score system.
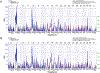
Results: Cardiomyocyte size varied significantly among BXD strains. Interval mapping on cardiomyocyte size data showed a significant QTL on chromosome (Chr) 2 at 66- 73.5 Mb and a suggestive QTL on Chr 5 at 20.9-39.7 Mb. Further score system revealed a high QTL score for Xirp2 in Chr 2. Xirp2 encodes xin actin-binding repeat containing 2, which is highly expressed in cardiac tissue and associate with cardiomyopathy and heart failure. In Chr5 QTL, Nos3, encoding nitric oxide synthase 3, received the highest score, which is significantly correlated with cardiomyocyte size.
Conclusion: These results indicate that Xirp2 and Nos3 serve as novel candidate modifier genes for myocardial hypertrophy in HCM. These candidate genes will be validated in our future studies.
Keywords: Hypertrophic cardiomyopathy; Modifier genes; Murine genetic reference population; Myocyte hypertrophy.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures




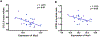
Similar articles
-
Echocardiography phenotyping in murine genetic reference population of BXD strains reveals significant QTLs associated with cardiac function and morphology.Physiol Genomics. 2023 Feb 1;55(2):51-66. doi: 10.1152/physiolgenomics.00120.2022. Epub 2022 Dec 19. Physiol Genomics. 2023. PMID: 36534598 Free PMC article.
-
Characterizing modifier genes of cardiac fibrosis phenotype in hypertrophic cardiomyopathy.Int J Cardiol. 2021 May 1;330:135-141. doi: 10.1016/j.ijcard.2021.01.047. Epub 2021 Jan 30. Int J Cardiol. 2021. PMID: 33529666 Free PMC article.
-
A Murine Hypertrophic Cardiomyopathy Model: The DBA/2J Strain.PLoS One. 2015 Aug 4;10(8):e0133132. doi: 10.1371/journal.pone.0133132. eCollection 2015. PLoS One. 2015. PMID: 26241864 Free PMC article.
-
Modifier genes for hypertrophic cardiomyopathy.Curr Opin Cardiol. 2002 May;17(3):242-52. doi: 10.1097/01.HCO.0000013803.40803.6A. Curr Opin Cardiol. 2002. PMID: 12015473 Free PMC article. Review.
-
Pathogenic Mechanisms of Hypertrophic Cardiomyopathy beyond Sarcomere Dysfunction.Int J Mol Sci. 2021 Aug 19;22(16):8933. doi: 10.3390/ijms22168933. Int J Mol Sci. 2021. PMID: 34445638 Free PMC article. Review.
Cited by
-
Unraveling the genetic blueprint of doxorubicin-induced cardiotoxicity through systems genetics approaches.Cardiooncology. 2025 Jun 3;11(1):53. doi: 10.1186/s40959-025-00349-y. Cardiooncology. 2025. PMID: 40462234 Free PMC article.
-
Cardiac copper content and its relationship with heart physiology: Insights based on quantitative genetic and functional analyses using BXD family mice.Front Cardiovasc Med. 2023 Feb 2;10:1089963. doi: 10.3389/fcvm.2023.1089963. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 36818345 Free PMC article.
-
Red ginseng prevents doxorubicin-induced cardiomyopathy by inhibiting cell death via activating the Nrf2 pathway.Cardiooncology. 2024 Jun 22;10(1):39. doi: 10.1186/s40959-024-00242-0. Cardiooncology. 2024. PMID: 38909271 Free PMC article.
-
Molecular Pathways and Animal Models of Cardiomyopathies.Adv Exp Med Biol. 2024;1441:991-1019. doi: 10.1007/978-3-031-44087-8_64. Adv Exp Med Biol. 2024. PMID: 38884766
-
Echocardiography phenotyping in murine genetic reference population of BXD strains reveals significant QTLs associated with cardiac function and morphology.Physiol Genomics. 2023 Feb 1;55(2):51-66. doi: 10.1152/physiolgenomics.00120.2022. Epub 2022 Dec 19. Physiol Genomics. 2023. PMID: 36534598 Free PMC article.
References
-
- Lopes LR, Rahman MS, Elliott PM, A systematic review and meta-analysis of genotype- phenotype associations in patients with hypertrophic cardiomyopathy caused by sarcomeric protein mutations, Heart 99(24) (2013) 1800–11. - PubMed
-
- Enriquez AD, Goldman ME, Management of hypertrophic cardiomyopathy, Ann Glob Health 80(1) (2014) 35–45. - PubMed
-
- Wigle ED, Rakowski H, Kimball BP, Williams WG, Hypertrophic cardiomyopathy. Clinical spectrum and treatment, Circulation 92(7) (1995) 1680–92. - PubMed
-
- Brugada R, Kelsey W, Lechin M, Zhao G, Yu QT, Zoghbi W, Quinones M, et al. Role of candidate modifier genes on the phenotypic expression of hypertrophy in patients with hypertrophic cardiomyopathy, J Investig Med 45(9) (1997) 542–51. - PubMed
-
- Chung MW, Tsoutsman T, Semsarian C, Hypertrophic cardiomyopathy: from gene defect to clinical disease, Cell Res 13(1) (2003) 9–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

