High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment
- PMID: 32470577
- PMCID: PMC7250083
- DOI: 10.1016/j.ijbiomac.2020.05.184
High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment
Abstract
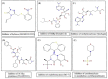
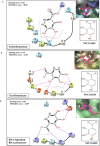
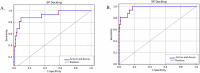
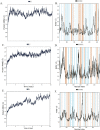
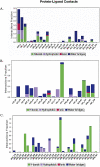
The present-day world is severely suffering from the recently emerged SARS-CoV-2. The lack of prescribed drugs for the deadly virus has stressed the likely need to identify novel inhibitors to alleviate and stop the pandemic. In the present high throughput virtual screening study, we used in silico techniques like receptor-ligand docking, Molecular dynamic (MD), and ADME properties to screen natural compounds. It has been documented that many natural compounds display antiviral activities, including anti-SARS-CoV effect. The present study deals with compounds of Natural Product Activity and Species Source (NPASS) database with known biological activity that probably impedes the activity of six essential enzymes of the virus. Promising drug-like compounds were identified, demonstrating better docking score and binding energy for each druggable targets. After an extensive screening analysis, three novel multi-target natural compounds were predicted to subdue the activity of three/more major drug targets simultaneously. Concerning the utility of natural compounds in the formulation of many therapies, we propose these compounds as excellent lead candidates for the development of therapeutic drugs against SARS-CoV-2.
Keywords: COVID-19; Molecular docking; Molecular dynamics; Muti-target; Natural compounds.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no conflicts of interest.
Figures



References
-
- Drosten C., Günther S., Preiser W., Van Der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

