Leptin Signaling Affects Survival and Chemoresistance of Estrogen Receptor Negative Breast Cancer
- PMID: 32471192
- PMCID: PMC7311967
- DOI: 10.3390/ijms21113794
Leptin Signaling Affects Survival and Chemoresistance of Estrogen Receptor Negative Breast Cancer
Abstract
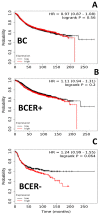
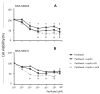
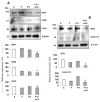
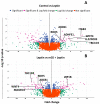
Estrogen-receptor-negative breast cancer (BCER-) is mainly treated with chemotherapeutics. Leptin signaling can influence BCER- progression, but its effects on patient survival and chemoresistance are not well understood. We hypothesize that leptin signaling decreases the survival of BCER- patients by, in part, inducing the expression of chemoresistance-related genes. The correlation of expression of leptin receptor (OBR), leptin-targeted genes (CDK8, NANOG, and RBP-Jk), and breast cancer (BC) patient survival was determined from The Cancer Genome Atlas (TCGA) mRNA data. Leptin-induced expression of proliferation and chemoresistance-related molecules was investigated in triple-negative BC (TNBC) cells that respond differently to chemotherapeutics. Leptin-induced gene expression in TNBC was analyzed by RNA-Seq. The specificity of leptin effects was assessed using OBR inhibitors (shRNA and peptides). The results show that OBR and leptin-targeted gene expression are associated with lower survival of BCER- patients. Importantly, the co-expression of these genes was also associated with chemotherapy failure. Leptin signaling increased the expression of tumorigenesis and chemoresistance-related genes (ABCB1, WNT4, ADHFE1, TBC1D3, LL22NC03, RDH5, and ITGB3) and impaired chemotherapeutic effects in TNBC cells. OBR inhibition re-sensitized TNBC to chemotherapeutics. In conclusion, the co-expression of OBR and leptin-targeted genes may be used as a predictor of survival and drug resistance of BCER- patients. Targeting OBR signaling could improve chemotherapeutic efficacy.
Keywords: chemoresistance; estrogen receptor negative breast cancer survival; leptin; leptin antagonist; obesity-related cancer.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures




References
-
- WHO Fact Sheets in Obesity and Overweight. [(accessed on 11 September 2019)]; Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight.
-
- For D., Benefit P. AACR Cancer Progress Report. AACR; Philadelphia, PA, USA: 2018.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

