Identification of a conserved var gene in different Plasmodium falciparum strains
- PMID: 32471507
- PMCID: PMC7260770
- DOI: 10.1186/s12936-020-03257-x
Identification of a conserved var gene in different Plasmodium falciparum strains
Abstract
Background: The multicopy var gene family of Plasmodium falciparum is of crucial importance for pathogenesis and antigenic variation. So far only var2csa, the var gene responsible for placental malaria, was found to be highly conserved among all P. falciparum strains. Here, a new conserved 3D7 var gene (PF3D7_0617400) is identified in several field isolates.
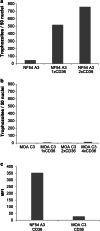
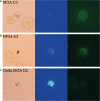
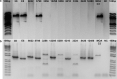
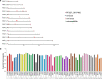
Methods: DNA sequencing, transcriptional analysis, Cluster of Differentiation (CD) 36-receptor binding, indirect immunofluorescence with PF3D7_0617400-antibodies and quantification of surface reactivity against semi-immune sera were used to characterize an NF54 clone and a Gabonese field isolate clone (MOA C3) transcribing the gene. A population of 714 whole genome sequenced parasites was analysed to characterize the conservation of the locus in African and Asian isolates. The genetic diversity of two var2csa fragments was compared with the genetic diversity of 57 microsatellites fragments in field isolates.
Results: PFGA01_060022400 was identified in a Gabonese parasite isolate (MOA) from a chronic infection and found to be 99% identical with PF3D7_0617400 of the 3D7 genome strain. Transcriptional analysis and immunofluorescence showed expression of the gene in an NF54 and a MOA clone but CD36 binding assays and surface reactivity to semi-immune sera differed markedly in the two clones. Long-read Pacific bioscience whole genome sequencing showed that PFGA01_060022400 is located in the internal cluster of chromosome 6. The full length PFGA01_060022400 was detected in 36 of 714 P. falciparum isolates and 500 bp fragments were identified in more than 100 isolates. var2csa was in parts highly conserved (He = 0) but in other parts as variable (He = 0.86) as the 57 microsatellites markers (He = 0.8).
Conclusions: Individual var gene sequences exhibit conservation in the global parasite population suggesting that purifying selection may limit overall genetic diversity of some var genes. Notably, field and laboratory isolates expressing the same var gene exhibit markedly different phenotypes.
Keywords: Genetic diversity; Malaria; Microsatellites; PfEMP1; Recombination; VSA; rifin; stevor; var genes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Whole genome sequencing and microsatellite analysis of the Plasmodium falciparum E5 NF54 strain show that the var, rifin and stevor gene families follow Mendelian inheritance.Malar J. 2018 Oct 22;17(1):376. doi: 10.1186/s12936-018-2503-2. Malar J. 2018. PMID: 30348135 Free PMC article.
-
CRISPR/Cas9 Genome Editing Reveals That the Intron Is Not Essential for var2csa Gene Activation or Silencing in Plasmodium falciparum.mBio. 2017 Jul 11;8(4):e00729-17. doi: 10.1128/mBio.00729-17. mBio. 2017. PMID: 28698275 Free PMC article.
-
Preferential transcription of conserved rif genes in two phenotypically distinct Plasmodium falciparum parasite lines.Int J Parasitol. 2009 May;39(6):655-64. doi: 10.1016/j.ijpara.2008.11.014. Epub 2008 Dec 30. Int J Parasitol. 2009. PMID: 19162031
-
In Vitro Variant Surface Antigen Expression in Plasmodium falciparum Parasites from a Semi-Immune Individual Is Not Correlated with Var Gene Transcription.PLoS One. 2016 Dec 1;11(12):e0166135. doi: 10.1371/journal.pone.0166135. eCollection 2016. PLoS One. 2016. PMID: 27907004 Free PMC article.
-
A systematic review on genetic diversity of var gene DBL1α domain from different geographical regions in Plasmodium falciparum isolates.Infect Genet Evol. 2021 Nov;95:105049. doi: 10.1016/j.meegid.2021.105049. Epub 2021 Aug 24. Infect Genet Evol. 2021. PMID: 34450294
Cited by
-
A novel computational pipeline for var gene expression augments the discovery of changes in the Plasmodium falciparum transcriptome during transition from in vivo to short-term in vitro culture.Elife. 2024 Jan 25;12:RP87726. doi: 10.7554/eLife.87726. Elife. 2024. PMID: 38270586 Free PMC article.
-
A paradoxical population structure of var DBLα types in Africa.bioRxiv [Preprint]. 2023 Nov 7:2023.11.05.565723. doi: 10.1101/2023.11.05.565723. bioRxiv. 2023. Update in: PLoS Pathog. 2025 Feb 04;21(2):e1012813. doi: 10.1371/journal.ppat.1012813. PMID: 37986738 Free PMC article. Updated. Preprint.
-
Plasmodium falciparum DNA repair dynamics reveal unique roles for TLS polymerases and PfRad51 in genome diversification.bioRxiv [Preprint]. 2025 Jun 21:2025.04.07.647301. doi: 10.1101/2025.04.07.647301. bioRxiv. 2025. PMID: 40667286 Free PMC article. Preprint.
-
Evasive mechanisms of human VSG and PfEMP1 antigens with link to Vaccine scenario: a review.J Parasit Dis. 2025 Mar;49(1):13-28. doi: 10.1007/s12639-024-01740-9. Epub 2024 Sep 24. J Parasit Dis. 2025. PMID: 39975623 Review.
-
Accuracy of diagnosis among clinical malaria patients: comparing microscopy, RDT and a highly sensitive quantitative PCR looking at the implications for submicroscopic infections.Malar J. 2023 Mar 4;22(1):76. doi: 10.1186/s12936-023-04506-5. Malar J. 2023. PMID: 36870966 Free PMC article.
References
-
- WHO. World Malaria Report 2016. Geneva, World Health Organization; 2016. http://www.who.int/malaria/publications/world-malaria-report-2016/report.... Accessed 17 Aug 2017.
-
- Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. - PubMed
-
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. - PubMed
-
- Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

