Introductions and early spread of SARS-CoV-2 in the New York City area
- PMID: 32471856
- PMCID: PMC7259823
- DOI: 10.1126/science.abc1917
Introductions and early spread of SARS-CoV-2 in the New York City area
Abstract
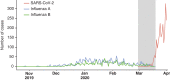
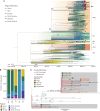
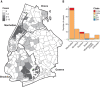
New York City (NYC) has emerged as one of the epicenters of the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. To identify the early transmission events underlying the rapid spread of the virus in the NYC metropolitan area, we sequenced the virus that causes coronavirus disease 2019 (COVID-19) in patients seeking care at the Mount Sinai Health System. Phylogenetic analysis of 84 distinct SARS-CoV-2 genomes indicates multiple, independent, but isolated introductions mainly from Europe and other parts of the United States. Moreover, we found evidence for community transmission of SARS-CoV-2 as suggested by clusters of related viruses found in patients living in different neighborhoods of the city.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures
References
-
- Executive Office of the President, Proclamation No. 9984, 85 F.R. 6709 (2020).
-
- Executive Office of the President, Proclamation No. 9992, 85 F.R. 12855 (2020).
-
- Executive Office of the President, Proclamation No. 9993, 85 F.R. 15045 (2020).
-
- Executive Office of the President, Proclamation No. 9996, 85 F.R. 15341 (2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

