CheY1 and CheY2 of Azorhizobium caulinodans ORS571 Regulate Chemotaxis and Competitive Colonization with the Host Plant
- PMID: 32471918
- PMCID: PMC7376556
- DOI: 10.1128/AEM.00599-20
CheY1 and CheY2 of Azorhizobium caulinodans ORS571 Regulate Chemotaxis and Competitive Colonization with the Host Plant
Abstract
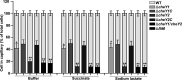
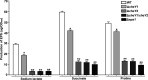
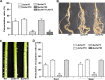
The genome of Azorhizobium caulinodans ORS571 encodes two chemotaxis response regulators: CheY1 and CheY2. cheY1 is located in a chemotaxis cluster (cheAWY1BR), while cheY2 is located 37 kb upstream of the cheAWY1BR cluster. To determine the contributions of CheY1 and CheY2, we compared the wild type (WT) and mutants in the free-living state and in symbiosis with the host Sesbania rostrata Swim plate tests and capillary assays revealed that both CheY1 and CheY2 play roles in chemotaxis, with CheY2 having a more prominent role than CheY1. In an analysis of the swimming paths of free-swimming cells, the ΔcheY1 mutant exhibited decreased frequency of direction reversal, whereas the ΔcheY2 mutant appeared to change direction much more frequently than the WT. Exopolysaccharide (EPS) production in the ΔcheY1 and ΔcheY2 mutants was lower than that in the WT, but the ΔcheY2 mutant had more obvious EPS defects that were similar to those of the ΔcheY1 ΔcheY2 and Δeps1 mutants. During symbiosis, the levels of competitiveness for root colonization and nodule occupation of ΔcheY1 and ΔcheY2 mutants were impaired compared to those of the WT. Moreover, the competitive colonization ability of the ΔcheY2 mutant was severely impaired compared to that of the ΔcheY1 mutant. Taken together, the ΔcheY2 phenotypes are more severe than the ΔcheY1 phenotype in free-living and symbiotic states, and that of the double mutant resembles the ΔcheY2 single-mutant phenotype. These defects of ΔcheY1 and ΔcheY2 mutants were restored to the WT phenotype by complementation. These results suggest that there are different regulatory mechanisms of CheY1 and CheY2 and that CheY2 is a key chemotaxis regulator under free-living and symbiosis conditions.IMPORTANCEAzorhizobium caulinodans ORS571 is a motile soil bacterium that has the dual capacity to fix nitrogen both under free-living conditions and in symbiosis with Sesbania rostrata, forming nitrogen-fixing root and stem nodules. Bacterial chemotaxis to chemoattractants derived from host roots promotes infection and subsequent nodule formation by directing rhizobia to appropriate sites of infection. In this work, we identified and demonstrated that CheY2, a chemotactic response regulator encoded by a gene outside the chemotaxis cluster, is required for chemotaxis and multiple other cell phenotypes. CheY1, encoded by a gene in the chemotaxis cluster, also plays a role in chemotaxis. Two response regulators mediate bacterial chemotaxis and motility in different ways. This work extends the understanding of the role of multiple response regulators in Gram-negative bacteria.
Keywords: Azorhizobium caulinodans; chemotaxis; response regulator CheY; symbiosis.
Copyright © 2020 American Society for Microbiology.
Figures
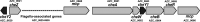


Similar articles
-
Azorhizobium caulinodans Chemotaxis Is Controlled by an Unusual Phosphorelay Network.J Bacteriol. 2022 Feb 15;204(2):e0052721. doi: 10.1128/JB.00527-21. Epub 2021 Nov 29. J Bacteriol. 2022. PMID: 34843377 Free PMC article.
-
A cheZ-Like Gene in Azorhizobium caulinodans Is a Key Gene in the Control of Chemotaxis and Colonization of the Host Plant.Appl Environ Microbiol. 2018 Jan 17;84(3):e01827-17. doi: 10.1128/AEM.01827-17. Print 2018 Feb 1. Appl Environ Microbiol. 2018. PMID: 29150498 Free PMC article.
-
A Chemotaxis Receptor Modulates Nodulation during the Azorhizobium caulinodans-Sesbania rostrata Symbiosis.Appl Environ Microbiol. 2016 May 16;82(11):3174-84. doi: 10.1128/AEM.00230-16. Print 2016 Jun 1. Appl Environ Microbiol. 2016. PMID: 26994081 Free PMC article.
-
Sensory transduction to the flagellar motor of Sinorhizobium meliloti.J Mol Microbiol Biotechnol. 2002 May;4(3):183-6. J Mol Microbiol Biotechnol. 2002. PMID: 11931544 Review.
-
Rhizobial Chemotaxis and Motility Systems at Work in the Soil.Front Plant Sci. 2021 Aug 27;12:725338. doi: 10.3389/fpls.2021.725338. eCollection 2021. Front Plant Sci. 2021. PMID: 34512702 Free PMC article. Review.
Cited by
-
The Divergent Key Residues of Two Agrobacterium fabrum (tumefaciens) CheY Paralogs Play a Key Role in Distinguishing Their Functions.Microorganisms. 2021 May 24;9(6):1134. doi: 10.3390/microorganisms9061134. Microorganisms. 2021. PMID: 34074050 Free PMC article.
-
The FtcR-Like Protein ActR in Azorhizobium caulinodans ORS571 Is Involved in Bacterial Motility and Symbiosis With the Host Plant.Front Microbiol. 2021 Nov 19;12:744268. doi: 10.3389/fmicb.2021.744268. eCollection 2021. Front Microbiol. 2021. PMID: 34867860 Free PMC article.
-
Rhizobium determinants of rhizosphere persistence and root colonization.ISME J. 2024 Jan 8;18(1):wrae072. doi: 10.1093/ismejo/wrae072. ISME J. 2024. PMID: 38690786 Free PMC article.
-
Protein Residues and a Novel Motif Involved in the Cellular Localization of CheZ in Azorhizobium caulinodans ORS571.Front Microbiol. 2020 Dec 7;11:585140. doi: 10.3389/fmicb.2020.585140. eCollection 2020. Front Microbiol. 2020. PMID: 33365019 Free PMC article.
-
Deciphering bacterial mechanisms of root colonization.Environ Microbiol Rep. 2021 Aug;13(4):428-444. doi: 10.1111/1758-2229.12934. Epub 2021 Feb 15. Environ Microbiol Rep. 2021. PMID: 33538402 Free PMC article. Review.
References
-
- Amsler CD, Matsumura P. 1995. Chemotactic signal transduction in Escherichia coli and Salmonella typhimurium, p 89–103. In Hoch J, Silhavy T (ed), Two-component signal transduction. ASM Press, Washington, DC.
-
- Stock JB, Surette MG. 1996. Chemotaxis, p 1103–1129. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC.
-
- Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE Jr. 1989. Identification of the site of phosphorylation of the chemotaxis response regulator CheY. J Biol Chem 264:21770–21778. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

