ISG15 and ISGylation is required for pancreatic cancer stem cell mitophagy and metabolic plasticity
- PMID: 32472071
- PMCID: PMC7260233
- DOI: 10.1038/s41467-020-16395-2
ISG15 and ISGylation is required for pancreatic cancer stem cell mitophagy and metabolic plasticity
Abstract
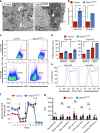
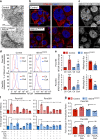
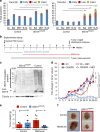
Pancreatic cancer stem cells (PaCSCs) drive pancreatic cancer tumorigenesis, chemoresistance and metastasis. While eliminating this subpopulation of cells would theoretically result in tumor eradication, PaCSCs are extremely plastic and can successfully adapt to targeted therapies. In this study, we demonstrate that PaCSCs increase expression of interferon-stimulated gene 15 (ISG15) and protein ISGylation, which are essential for maintaining their metabolic plasticity. CRISPR-mediated ISG15 genomic editing reduces overall ISGylation, impairing PaCSCs self-renewal and their in vivo tumorigenic capacity. At the molecular level, ISG15 loss results in decreased mitochondrial ISGylation concomitant with increased accumulation of dysfunctional mitochondria, reduced oxidative phosphorylation (OXPHOS) and impaired mitophagy. Importantly, disruption in mitochondrial metabolism affects PaCSC metabolic plasticity, making them susceptible to prolonged inhibition with metformin in vivo. Thus, ISGylation is critical for optimal and efficient OXPHOS by ensuring the recycling of dysfunctional mitochondria, and when absent, a dysregulation in mitophagy occurs that negatively impacts PaCSC stemness.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. - PubMed
-
- Hidalgo M, et al. Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology. 2015;15:8–18. - PubMed
-
- Ko AH. Progress in the treatment of metastatic pancreatic cancer and the search for next opportunities. J. Clin. Oncol. 2015;33:1779–1786. - PubMed
-
- de Sousa e Melo F, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

