SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective
- PMID: 32473976
- PMCID: PMC7256558
- DOI: 10.1016/j.meegid.2020.104384
SARS-CoV-2 and COVID-19: A genetic, epidemiological, and evolutionary perspective
Abstract
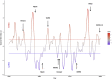
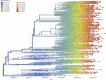
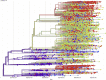
In less than five months, COVID-19 has spread from a small focus in Wuhan, China, to more than 5 million people in almost every country in the world, dominating the concern of most governments and public health systems. The social and political distresses caused by this epidemic will certainly impact our world for a long time to come. Here, we synthesize lessons from a range of scientific perspectives rooted in epidemiology, virology, genetics, ecology and evolutionary biology so as to provide perspective on how this pandemic started, how it is developing, and how best we can stop it.
Keywords: Coevolution; Coronavirus; Host susceptibility; Immune system; Pandemics; Phylodynamics.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

