Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type
- PMID: 32474553
- PMCID: PMC7530441
- DOI: 10.4103/ijmr.IJMR_1125_20
Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type
Abstract
Background & objectives: SARS-CoV-2 (Severe acute respiratory syndrome coronavirus-2) is evolving with the progression of the pandemic. This study was aimed to investigate the diversity and evolution of the coronavirus SARS-CoV-2 with progression of the pandemic over time and to identify similarities and differences of viral diversity and evolution across geographical regions (countries).
Methods: Publicly available data on type definitions based on whole-genome sequences of the SARS-CoV-2 sampled during December and March 2020 from 3636 infected patients spread over 55 countries were collected. Phylodynamic analyses were performed and the temporal and spatial evolution of the virus was examined.
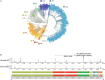
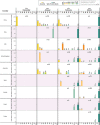
Results: It was found that (i) temporal variation in frequencies of types of the coronavirus was significant; ancestral viruses of type O were replaced by evolved viruses belonging to type A2a; (ii) spatial variation was not significant; with the spread of SARS-CoV-2, the dominant virus was the A2a type virus in every geographical region; (iii) within a geographical region, there was significant micro-level variation in the frequencies of the different viral types, and (iv) the evolved coronavirus of type A2a swept rapidly across all continents.
Interpretation & conclusions: SARS-CoV-2 belonging to the A2a type possesses a non-synomymous variant (D614G) that possibly eases the entry of the virus into the lung cells of the host. This may be the reason why the A2a type has an advantage to infect and survive and as a result has rapidly swept all geographical regions. Therefore, large-scale sequencing of coronavirus genomes and, as required, of host genomes should be undertaken in India to identify regional and ethnic variation in viral composition and its interaction with host genomes. Further, careful collection of clinical and immunological data of the host can provide deep learning in relation to infection and transmission of the types of coronavirus genomes.
Keywords: Host genome interaction - phylogeny - RNA sequence - SARS-CoV-2 - viral type coronavirus.
Conflict of interest statement
None
Figures


References
-
- World Health Organization. Update 49 - SARS case fatality ratio, incubation period. WHO; 2003. [accessed on April 9, 2020]. Available from: https://wwwwhoint/csr/sars/archive/2003_05_07a/en/
-
- World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV) WHO; 2020. [accessed on April 9, 2020]. Available from: https://wwwwhoint/emergencies/mers-cov/en/">https://wwwwhoint/emergencie...
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
