PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics
- PMID: 32475992
- PMCID: PMC7319888
- DOI: 10.1038/s41388-020-1336-y
PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics
Abstract
Using PROteolysis TArgeting Chimeras (PROTACs) to degrade proteins that are important for tumorigenesis has emerged as a potential therapeutic strategy for cancer. PROTACs are heterobifunctional molecules consisting of one ligand for binding to a protein of interest (POI) and another to an E3 ubiquitin (E3) ligase, connected via a linker. PROTACs recruit the E3 ligase to the POI and cause proximity-induced ubiquitination and degradation of the POI by the ubiquitin-proteasome system (UPS). PROTACs have been developed to degrade a variety of cancer targets with unprecedented efficacy against a multitude of tumor types. To date, most of the PROTACs developed have utilized ligands to recruit E3 ligases that are ubiquitously expressed in both tumor and normal tissues. These PROTACs can cause on-target toxicities if the POIs are not tumor-specific. Therefore, identifying and recruiting the E3 ligases that are enriched in tumors with minimal expression in normal tissues holds the potential to develop tumor-specific/selective PROTACs. In this review, we will discuss the potential of PROTACs to become anticancer therapeutics, chemical and bioinformatics approaches for PROTAC design, and safety concerns with a special focus on the development of tumor-specific/selective PROTACs. In addition, the identification of tumor types in terms of solid versus hematological malignancies that can be best targeted with PROTAC approach will be briefly discussed.
Conflict of interest statement
Figures
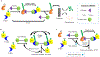

References
-
- Zhao Q, Ren C, Liu L, Chen J, Shao Y, Sun N et al. Discovery of SIAIS178 as an Effective BCR-ABL Degrader by Recruiting Von Hippel-Lindau (VHL) E3 Ubiquitin Ligase. J Med Chem 2019; 62: 9281–9298. - PubMed
-
- Gonzalez TL, Hancock M, Sun S, Gersch CL, Larios JM, David W et al. Targeted degradation of activating estrogen receptor α ligand-binding domain mutations in human breast cancer. Breast Cancer Res Treat 2020; 180: 611–622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

