Serum proteomics analysis of feline mammary carcinoma based on label-free and PRM techniques
- PMID: 32476319
- PMCID: PMC7263907
- DOI: 10.4142/jvs.2020.21.e45
Serum proteomics analysis of feline mammary carcinoma based on label-free and PRM techniques
Abstract
Background: Feline mammary carcinoma is the third most common cancer that affects female cats.
Objectives: The purpose of this study was to screen differential serum proteins in feline and clarify the relationship between them and the occurrence of feline mammary carcinoma.
Methods: Chinese pastoral cats were used as experimental animals. Six serum samples from cats with mammary carcinoma (group T) and six serum samples from healthy cats (group C) were selected. Differential protein analysis was performed using a Label-free technique, while parallel reaction monitoring (PRM) was performed to verify the screened differential proteins.
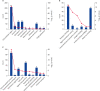
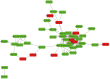
Results: A total of 82 differential proteins were detected between group T and group C, of which 55 proteins were down regulated and 27 proteins were up regulated. Apolipoprotein A-I, Apolipoprotein A-II (ApoA-II), Apolipoprotein B (ApoB), Apolipoprotein C-III (ApoC-III), coagulation factor V, coagulation factor X, C1q, albumen (ALB) were all associated with the occurrence of feline mammary carcinoma. Differential proteins were involved in a total of 40 signaling pathways, among which the metabolic pathways associated with feline mammary carcinoma were the complement and coagulation cascade and cholesterol metabolism. According to the Label-free results, ApoB, ApoC-III, ApoA-II, FN1, an uncharacterized protein, and ALB were selected for PRM target verification. The results were consistent with the trend of the label-free.
Conclusions: This experimen is the first to confirm ApoA-II and ApoB maybe new feline mammary carcinoma biomarkers and to analyze their mechanisms in the development of such carcinoma in feline.
Keywords: Feline mammary carcinoma; PRM; label-free; proteomics.
© 2020 The Korean Society of Veterinary Science.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures


Similar articles
-
Analysis of serum peptidome profiles of non-metastatic and metastatic feline mammary carcinoma using liquid chromatography-tandem mass spectrometry.BMC Vet Res. 2024 Jun 29;20(1):280. doi: 10.1186/s12917-024-04148-y. BMC Vet Res. 2024. PMID: 38951817 Free PMC article.
-
The implication of autoantibodies in early diagnosis and monitoring of plasmonic photothermal therapy in the treatment of feline mammary carcinoma.Sci Rep. 2021 May 17;11(1):10441. doi: 10.1038/s41598-021-89894-x. Sci Rep. 2021. PMID: 34001936 Free PMC article.
-
Prognostic value of histologic grading for feline mammary carcinoma: a retrospective survival analysis.Vet Pathol. 2015 Mar;52(2):238-49. doi: 10.1177/0300985814543198. Epub 2014 Jul 24. Vet Pathol. 2015. PMID: 25060990
-
Skeletal metastasis in feline mammary carcinoma: case report and literature review.J Am Anim Hosp Assoc. 1998 Mar-Apr;34(2):103-8. doi: 10.5326/15473317-34-2-103. J Am Anim Hosp Assoc. 1998. PMID: 9507421 Review.
-
Carcinocythemia: First report in a cat and literature review.Vet Clin Pathol. 2018 Mar;47(1):142-145. doi: 10.1111/vcp.12565. Epub 2018 Jan 23. Vet Clin Pathol. 2018. PMID: 29360147 Review.
Cited by
-
Analysis of serum peptidome profiles of non-metastatic and metastatic feline mammary carcinoma using liquid chromatography-tandem mass spectrometry.BMC Vet Res. 2024 Jun 29;20(1):280. doi: 10.1186/s12917-024-04148-y. BMC Vet Res. 2024. PMID: 38951817 Free PMC article.
-
Feline Mammary Tumors: A Comprehensive Review of Histological Classification Schemes, Grading Systems, and Prognostic Factors.Vet Sci. 2025 Aug 5;12(8):736. doi: 10.3390/vetsci12080736. Vet Sci. 2025. PMID: 40872687 Free PMC article. Review.
-
Serum proteome profiles in cats with chronic enteropathies.J Vet Intern Med. 2023 Jul-Aug;37(4):1358-1367. doi: 10.1111/jvim.16743. Epub 2023 Jun 6. J Vet Intern Med. 2023. PMID: 37279179 Free PMC article.
-
Label-free quantitative proteomics analysis of jujube (Ziziphus jujuba Mill.) during different growth stages.RSC Adv. 2021 Jun 22;11(36):22106-22119. doi: 10.1039/d1ra02989d. eCollection 2021 Jun 21. RSC Adv. 2021. PMID: 35480818 Free PMC article.
-
Presence of immune factors in freshwater mussel (Hyriopsis cumingii) entails autologous serum an essential component in the culture of mantle cells.Front Immunol. 2023 May 5;14:1173184. doi: 10.3389/fimmu.2023.1173184. eCollection 2023. Front Immunol. 2023. PMID: 37215128 Free PMC article.
References
-
- Zappulli V, Rasotto R, Caliari D, Mainenti M, Peña L, Goldschmidt MH, Kiupel M. Prognostic evaluation of feline mammary carcinomas: a review of the literature. Vet Pathol. 2015;52(1):46–60. - PubMed
-
- Weijer K, Hart A A M. Prognostic factors in feline mammary carcinoma. J Natl Cancer Inst. 1983;70(4):709–716. - PubMed
-
- Eckert MA, Coscia F, Chryplewicz A, Chang JW, Hernandez KM, Pan S, Tienda SM, Nahotko DA, Li G, Blaženović I, Lastra RR, Curtis M, Yamada SD, Perets R, McGregor SM, Andrade J, Fiehn O, Moellering RE, Mann M, Lengyel E. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569(7758):723–728. - PMC - PubMed
-
- Marx H, Minogue CE, Jayaraman D, Richards AL, Kwiecien NW, Siahpirani AF, Rajasekar S, Maeda J, Garcia K, Del Valle-Echevarria AR, Volkening JD, Westphall MS, Roy S, Sussman MR, Ané JM, Coon JJ. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti . Nat Biotechnol. 2016;34(11):1198–1205. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- LBH-Z18257/Heilongjiang Provincial Postdoctoral Science Foundation/China
- TDJH201903/Heilongjiang Bayi Agricultural University Postdoctoral science foundation/China
- 2016YFD0501008/National key research and development program of China/China
- YJSCX2018-Y23/Heilongjiang Bayi Agricultural University Postgraduate research innovation funding program/China
LinkOut - more resources
Full Text Sources
Miscellaneous

