Genomic and Phenotypic Heterogeneity of Clinical Isolates of the Human Pathogens Aspergillus fumigatus, Aspergillus lentulus, and Aspergillus fumigatiaffinis
- PMID: 32477406
- PMCID: PMC7236307
- DOI: 10.3389/fgene.2020.00459
Genomic and Phenotypic Heterogeneity of Clinical Isolates of the Human Pathogens Aspergillus fumigatus, Aspergillus lentulus, and Aspergillus fumigatiaffinis
Abstract
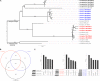
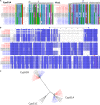
Fungal pathogens are a global threat to human health. For example, fungi from the genus Aspergillus cause a spectrum of diseases collectively known as aspergillosis. Most of the >200,000 life-threatening aspergillosis infections per year worldwide are caused by Aspergillus fumigatus. Recently, molecular typing techniques have revealed that aspergillosis can also be caused by organisms that are phenotypically similar to A. fumigatus but genetically distinct, such as Aspergillus lentulus and Aspergillus fumigatiaffinis. Importantly, some of these so-called cryptic species are thought to exhibit different virulence and drug susceptibility profiles than A. fumigatus, however, our understanding of their biology and pathogenic potential has been stymied by the lack of genome sequences and phenotypic profiling of multiple clinical strains. To fill this gap, we phenotypically characterized the virulence and drug susceptibility of 15 clinical strains of A. fumigatus, A. lentulus, and A. fumigatiaffinis from Spain and sequenced their genomes. We found heterogeneity in drug susceptibility across species and strains. We further found heterogeneity in virulence within each species but no significant differences in the virulence profiles between the three species. Genes known to influence drug susceptibility (cyp51A and fks1) vary in paralog number and sequence among these species and strains and correlate with differences in drug susceptibility. Similarly, genes known to be important for virulence in A. fumigatus showed variability in number of paralogs across strains and across species. Characterization of the genomic similarities and differences of clinical strains of A. lentulus, A. fumigatiaffinis, and A. fumigatus that vary in disease-relevant traits will advance our understanding of the variance in pathogenicity between Aspergillus species and strains that are collectively responsible for the vast majority of aspergillosis infections in humans.
Keywords: Aspergillus; antifungal drug susceptibility; cryptic species; drug resistance; genetic determinants of virulence; genomics; strain heterogeneity; virulence.
Copyright © 2020 dos Santos, Steenwyk, Rivero-Menendez, Mead, Silva, Bastos, Alastruey-Izquierdo, Goldman and Rokas.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

