Revisiting Salisapiliaceae
- PMID: 32478314
- PMCID: PMC7252517
- DOI: 10.3114/fuse.2019.03.10
Revisiting Salisapiliaceae
Abstract
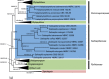
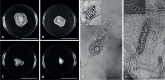
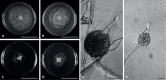
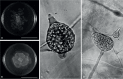
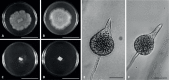
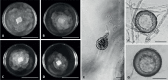
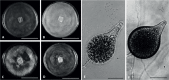
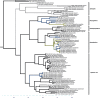
Of the diverse lineages of the Phylum Oomycota, saprotrophic oomycetes from the salt marsh and mangrove habitats are still understudied, despite their ecological importance. Salisapiliaceae, a monophyletic and monogeneric taxon of the marine and estuarine oomycetes, was introduced to accommodate species with a protruding hyaline apical plug, small hyphal diameter and lack of vesicle formation during zoospore release. At the time of description of Salisapilia, only few species of Halophytophthora, an ecologically similar, phylogenetically heterogeneous genus from which Salisapilia was segregated, were included. In this study, a revision of the genus Salisapilia is presented, and five new combinations (S. bahamensis, S. elongata, S. epistomia, S. masteri, and S. mycoparasitica) and one new species (S. coffeyi) are proposed. Further, the species description of S. nakagirii is emended for some exceptional morphological and developmental characteristics. A key to the genus Salisapilia is provided and its generic circumscription and character evolution in cultivable Peronosporales are discussed.
Keywords: Estuarine oomycetes; Halophytophthora; Salisapilia; mangroves; new taxa; phylogeny.
© 2019 Westerdijk Fungal Biodiversity Institute.
Figures





References
-
- Anastasiou CJ, Churchland LM. (1969). Fungi on decaying leaves in marine habitats. Canadian Journal of Botany 47: 251–257.
-
- Bachofer M. (2004). Molekularbiologische Populationsstudien an Plasmopara halstedii, dem Falschen Mehltau der Sonnenblume. PhD thesis, University of Hohenheim, Germany.
-
- Bala K, Robideau GP, Lévesque CA, et al. (2010). Phytopythium Abad, de Cock, Bala, Robideau, Lodhi and Lévesque, gen. nov. and Phytopythium sindhum Lodhi, Shahzad and Lévesque, sp. nov. Persoonia 24: 136–137.
-
- Beakes GW, Thines M. (2017). Hyphochytriomycota and Oomycota. In: Handbook of the Protists (Archibald JM, Simpson AGB, Slamovits C, eds). Springer International Publishing, Germany: 435–505.
-
- Bennett RM, Dedeles GR, Thines M. (2017a). Phytophthora elongata (Peronosporaceae) is present as an estuarine species in Philippine mangroves. Mycosphere 8: 959–967.
LinkOut - more resources
Full Text Sources
