Retrocopying expands the functional repertoire of APOBEC3 antiviral proteins in primates
- PMID: 32479260
- PMCID: PMC7263822
- DOI: 10.7554/eLife.58436
Retrocopying expands the functional repertoire of APOBEC3 antiviral proteins in primates
Abstract
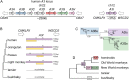
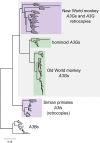
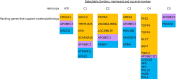
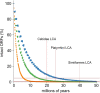
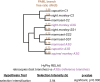
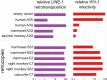
Host-virus arms races are inherently asymmetric; viruses evolve much more rapidly than host genomes. Thus, there is high interest in discovering mechanisms by which host genomes keep pace with rapidly evolving viruses. One family of restriction factors, the APOBEC3 (A3) cytidine deaminases, has undergone positive selection and expansion via segmental gene duplication and recombination. Here, we show that new copies of A3 genes have also been created in primates by reverse transcriptase-encoding elements like LINE-1 or endogenous retroviruses via a process termed retrocopying. First, we discovered that all simian primate genomes retain the remnants of an ancient A3 retrocopy: A3I. Furthermore, we found that some New World monkeys encode up to ten additional APOBEC3G (A3G) retrocopies. Some of these A3G retrocopies are transcribed in a variety of tissues and able to restrict retroviruses. Our findings suggest that host genomes co-opt retroelement activity in the germline to create new host restriction factors as another means to keep pace with the rapid evolution of viruses. (163).
Keywords: APOBEC3; evolutionary biology; genome defense; genome evolution; infectious disease; microbiology; restriction factors; retrogenes; transposable elements.
© 2020, Yang et al.
Conflict of interest statement
LY, ME, HM, RM No competing interests declared
Figures





Similar articles
-
Retroviruses drive the rapid evolution of mammalian APOBEC3 genes.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):610-618. doi: 10.1073/pnas.1914183116. Epub 2019 Dec 16. Proc Natl Acad Sci U S A. 2020. PMID: 31843890 Free PMC article.
-
Mutational resilience of antiviral restriction favors primate TRIM5α in host-virus evolutionary arms races.Elife. 2020 Sep 15;9:e59988. doi: 10.7554/eLife.59988. Elife. 2020. PMID: 32930662 Free PMC article.
-
Evidence for restriction of ancient primate gammaretroviruses by APOBEC3 but not TRIM5alpha proteins.PLoS Pathog. 2008 Oct;4(10):e1000181. doi: 10.1371/journal.ppat.1000181. Epub 2008 Oct 17. PLoS Pathog. 2008. PMID: 18927623 Free PMC article.
-
The Battle between Retroviruses and APOBEC3 Genes: Its Past and Present.Viruses. 2021 Jan 17;13(1):124. doi: 10.3390/v13010124. Viruses. 2021. PMID: 33477360 Free PMC article. Review.
-
[Molecular evolution of physiologically functioning anti-retroviral APOBEC3 deaminases].Uirusu. 2012 Jun;62(1):27-38. doi: 10.2222/jsv.62.27. Uirusu. 2012. PMID: 23189822 Review. Japanese.
Cited by
-
Rapid Evolution of HERC6 and Duplication of a Chimeric HERC5/6 Gene in Rodents and Bats Suggest an Overlooked Role of HERCs in Mammalian Immunity.Front Immunol. 2020 Dec 18;11:605270. doi: 10.3389/fimmu.2020.605270. eCollection 2020. Front Immunol. 2020. PMID: 33391270 Free PMC article.
-
Variability in Codon Usage in Coronaviruses Is Mainly Driven by Mutational Bias and Selective Constraints on CpG Dinucleotide.Viruses. 2021 Sep 10;13(9):1800. doi: 10.3390/v13091800. Viruses. 2021. PMID: 34578381 Free PMC article.
-
Highly-potent, synthetic APOBEC3s restrict HIV-1 through deamination-independent mechanisms.PLoS Pathog. 2021 Jun 25;17(6):e1009523. doi: 10.1371/journal.ppat.1009523. eCollection 2021 Jun. PLoS Pathog. 2021. PMID: 34170969 Free PMC article.
-
APOBEC3 deaminase editing in mpox virus as evidence for sustained human transmission since at least 2016.Science. 2023 Nov 3;382(6670):595-600. doi: 10.1126/science.adg8116. Epub 2023 Nov 2. Science. 2023. PMID: 37917680 Free PMC article.
-
Factors Regulating the Activity of LINE1 Retrotransposons.Genes (Basel). 2021 Sep 30;12(10):1562. doi: 10.3390/genes12101562. Genes (Basel). 2021. PMID: 34680956 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

