Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs
- PMID: 32479594
- PMCID: PMC7401813
- DOI: 10.1083/jcb.201907058
Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs
Abstract
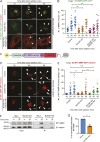
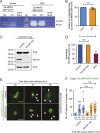
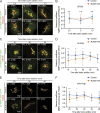
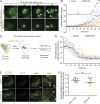
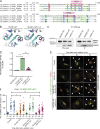
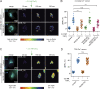
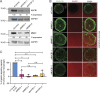
Matrix metalloproteinases (MMPs) degrade several ECM components and are crucial modulators of cell invasion and tissue organization. Although much has been reported about their function in remodeling ECM in health and disease, their trafficking across the Golgi apparatus remains poorly understood. Here we report that the cis-Golgi protein nucleobindin-1 (NUCB1) is critical for MMP2 and MT1-MMP trafficking along the Golgi apparatus. This process is Ca2+-dependent and is required for invasive MDA-MB-231 cell migration as well as for gelatin degradation in primary human macrophages. Our findings emphasize the importance of NUCB1 as an essential component of MMP transport and its overall impact on ECM remodeling.
© 2020 Pacheco-Fernandez et al.
Figures


Similar articles
-
RAB2A controls MT1-MMP endocytic and E-cadherin polarized Golgi trafficking to promote invasive breast cancer programs.EMBO Rep. 2016 Jul;17(7):1061-80. doi: 10.15252/embr.201642032. Epub 2016 Jun 2. EMBO Rep. 2016. PMID: 27255086 Free PMC article.
-
Less is more: low expression of MT1-MMP is optimal to promote migration and tumourigenesis of breast cancer cells.Mol Cancer. 2016 Oct 18;15(1):65. doi: 10.1186/s12943-016-0547-x. Mol Cancer. 2016. PMID: 27756325 Free PMC article.
-
VAMP3, syntaxin-13 and SNAP23 are involved in secretion of matrix metalloproteinases, degradation of the extracellular matrix and cell invasion.J Cell Sci. 2009 Nov 15;122(Pt 22):4089-98. doi: 10.1242/jcs.052761. J Cell Sci. 2009. PMID: 19910495
-
The membrane tethered matrix metalloproteinase MT1-MMP at the forefront of melanoma cell invasion and metastasis.Pharmacol Res. 2016 Sep;111:17-22. doi: 10.1016/j.phrs.2016.05.019. Epub 2016 May 21. Pharmacol Res. 2016. PMID: 27221755 Review.
-
Novel Roles of MT1-MMP and MMP-2: Beyond the Extracellular Milieu.Int J Mol Sci. 2022 Aug 23;23(17):9513. doi: 10.3390/ijms23179513. Int J Mol Sci. 2022. PMID: 36076910 Free PMC article. Review.
Cited by
-
TEM1 up-regulates MMP-2 and promotes ECM remodeling for facilitating invasion and migration of uterine sarcoma.Discov Oncol. 2023 Jan 13;14(1):5. doi: 10.1007/s12672-023-00613-6. Discov Oncol. 2023. PMID: 36639546 Free PMC article.
-
Meta-Analysis of MS-Based Proteomics Studies Indicates Interferon Regulatory Factor 4 and Nucleobindin1 as Potential Prognostic and Drug Resistance Biomarkers in Diffuse Large B Cell Lymphoma.Cells. 2023 Jan 3;12(1):196. doi: 10.3390/cells12010196. Cells. 2023. PMID: 36611989 Free PMC article. Review.
-
Ca2+ regulation of constitutive vesicle trafficking.Fac Rev. 2022 Mar 9;11:6. doi: 10.12703/r/11-6. eCollection 2022. Fac Rev. 2022. PMID: 35359486 Free PMC article. Review.
-
Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells.Bio Protoc. 2021 Mar 5;11(5):e3936. doi: 10.21769/BioProtoc.3936. eCollection 2021 Mar 5. Bio Protoc. 2021. PMID: 33796610 Free PMC article.
-
Deciphering the Kidney Matrisome: Identification and Quantification of Renal Extracellular Matrix Proteins in Healthy Mice.Int J Mol Sci. 2023 Feb 1;24(3):2827. doi: 10.3390/ijms24032827. Int J Mol Sci. 2023. PMID: 36769148 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

