Dynamics of the 4D genome during in vivo lineage specification and differentiation
- PMID: 32483172
- PMCID: PMC7264236
- DOI: 10.1038/s41467-020-16598-7
Dynamics of the 4D genome during in vivo lineage specification and differentiation
Abstract
Mammalian gene expression patterns are controlled by regulatory elements, which interact within topologically associating domains (TADs). The relationship between activation of regulatory elements, formation of structural chromatin interactions and gene expression during development is unclear. Here, we present Tiled-C, a low-input chromosome conformation capture (3C) technique. We use this approach to study chromatin architecture at high spatial and temporal resolution through in vivo mouse erythroid differentiation. Integrated analysis of chromatin accessibility and single-cell expression data shows that regulatory elements gradually become accessible within pre-existing TADs during early differentiation. This is followed by structural re-organization within the TAD and formation of specific contacts between enhancers and promoters. Our high-resolution data show that these enhancer-promoter interactions are not established prior to gene expression, but formed gradually during differentiation, concomitant with progressive upregulation of gene activity. Together, these results provide new insight into the close, interdependent relationship between chromatin architecture and gene regulation during development.
Conflict of interest statement
J.R.H. is founder and shareholder of Nucleome Therapeutics.
Figures
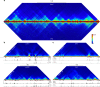
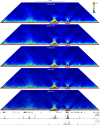
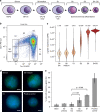
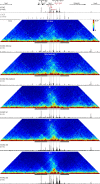

References
-
- Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019;20:437–455. - PubMed
-
- Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354. - PubMed
-
- Andrey G, Mundlos S. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017;144:3646–3658. - PubMed
-
- de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12009/1/MRC_/Medical Research Council/United Kingdom
- MR/N00969X/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/15/MRC_/Medical Research Council/United Kingdom
- MC_U137961145/MRC_/Medical Research Council/United Kingdom
- MC_U137961144/MRC_/Medical Research Council/United Kingdom
- MC_PC_15069/MRC_/Medical Research Council/United Kingdom
- G1000801/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/4/MRC_/Medical Research Council/United Kingdom
- 209181/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_12009/13/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/1/MRC_/Medical Research Council/United Kingdom
- R01 DK100915/DK/NIDDK NIH HHS/United States
- MC_UU_00016/12/MRC_/Medical Research Council/United Kingdom
- BB/R008655/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M025624/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UU_00016/14/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/4/MRC_/Medical Research Council/United Kingdom
- BB/J001694/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

