Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients
- PMID: 32483236
- PMCID: PMC7264175
- DOI: 10.1038/s41467-020-16638-2
Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients
Abstract
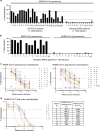
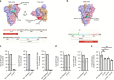
Given the ongoing SARS-CoV-2 pandemic, identification of immunogenic targets against the coronavirus spike glycoprotein will provide crucial advances towards the development of sensitive diagnostic tools and potential vaccine candidate targets. In this study, using pools of overlapping linear B-cell peptides, we report two IgG immunodominant regions on SARS-CoV-2 spike glycoprotein that are recognised by sera from COVID-19 convalescent patients. Notably, one is specific to SARS-CoV-2, which is located in close proximity to the receptor binding domain. The other region, which is localised at the fusion peptide, could potentially function as a pan-SARS target. Functionally, antibody depletion assays demonstrate that antibodies targeting these immunodominant regions significantly alter virus neutralisation capacities. Taken together, identification and validation of these neutralising B-cell epitopes will provide insights towards the design of diagnostics and vaccine candidates against this high priority coronavirus.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Cohen J, Normile D. New SARS-like virus in China triggers alarm. Science. 2020;367:234–235. - PubMed
-
- Hoffmann Markus, Kleine-Weber Hannah, Schroeder Simon, Krüger Nadine, Herrler Tanja, Erichsen Sandra, Schiergens Tobias S., Herrler Georg, Wu Nai-Huei, Nitsche Andreas, Müller Marcel A., Drosten Christian, Pöhlmann Stefan. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271-280.e8. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

