Mitochondrial genetic variation reveals phylogeographic structure and cryptic diversity in Trioza erytreae
- PMID: 32483282
- PMCID: PMC7264336
- DOI: 10.1038/s41598-020-65880-7
Mitochondrial genetic variation reveals phylogeographic structure and cryptic diversity in Trioza erytreae
Erratum in
-
Author Correction: Mitochondrial genetic variation reveals phylogeographic structure and cryptic diversity in Trioza erytreae.Sci Rep. 2020 Nov 13;10(1):20204. doi: 10.1038/s41598-020-76731-w. Sci Rep. 2020. PMID: 33188268 Free PMC article.
Abstract
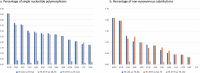
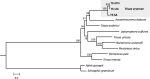
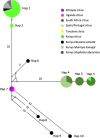
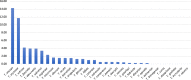
Trioza erytreae is the main vector for 'Candidatus Liberibacter africanus', the causative agent of African Citrus Greening disease. The insect is widespread in Africa, and has recently disseminated to Southwestern Europe. This study aimed at generating reference mitogenome sequences for T. erytreae, as a background for future genetic diversity surveys. Complete mitochondrial sequences of three specimens collected in Ethiopia, Uganda and South Africa were recovered using Ion Torrent technology. The mitogenomes of T. erytreae from Uganda and Ethiopia were highly similar, and distinct from that found in South Africa. The phylogeographic structure of T. erytreae was assessed using genetic clustering and pairwise distances, based on a dataset of public COI sequences recorded as T. erytreae. The dataset revealed ten haplotypes with strong phylogeographic structure in Africa and Europe. Three haplotypes found in Kenya on Clausena anisata belonged to pairs separated by distances as high as 11.2%, and were basal to all other sequences. These results indicate that not all sequences identified as T. erytreae belong to the same species, and that some degree of specificity with different plant hosts is likely to exist. This study provides new baseline information on the diversity of T. erytreae, with potential implications for the epidemiology of African Citrus Greening disease.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Food and Agriculture Organization of the United Nations. FAOSTAT; http://www.fao.org/faostat/en/#home Accessed November 11, 2019 (2019).
-
- Kilalo D, Olubayo F, Obukosia S, Shibairo S. Farmer management practices of citrus insect pests in Kenya. Afr. J. Hort. Sci. 2009;2:168–176.
-
- Nyambo, B. Integrated pest management plan. The Agricultural Sector Development Program - Republic of Tanzania, pp 57 (2009).
-
- Ekesi, S. Arthropod pest composition and farmers perceptions of pest and disease problems on citrus in Kenya. XII International Citrus Conference. pp 283 (2012).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

